Full-Length Recombinant hSP-D Binds and Inhibits SARS-CoV-2
- PMID: 34439781
- PMCID: PMC8393632
- DOI: 10.3390/biom11081114
Full-Length Recombinant hSP-D Binds and Inhibits SARS-CoV-2
Abstract
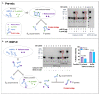
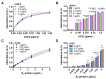
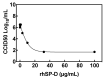
SARS-CoV-2 infection of host cells is driven by binding of the SARS-CoV-2 spike-(S)-protein to lung type II pneumocytes, followed by virus replication. Surfactant protein SP-D, member of the front-line immune defense of the lungs, binds glycosylated structures on invading pathogens such as viruses to induce their clearance from the lungs. The objective of this study is to measure the pulmonary SP-D levels in COVID-19 patients and demonstrate the activity of SP-D against SARS-CoV-2, opening the possibility of using SP-D as potential therapy for COVID-19 patients. Pulmonary SP-D concentrations were measured in bronchoalveolar lavage samples from patients with corona virus disease 2019 (COVID-19) by anti-SP-D ELISA. Binding assays were performed by ELISAs. Protein bridge and aggregation assays were performed by gel electrophoresis followed by silver staining and band densitometry. Viral replication was evaluated in vitro using epithelial Caco-2 cells. Results indicate that COVID-19 patients (n = 12) show decreased pulmonary levels of SP-D (median = 68.9 ng/mL) when compared to levels reported for healthy controls in literature. Binding assays demonstrate that SP-D binds the SARS-CoV-2 glycosylated spike-(S)-protein of different emerging clinical variants. Binding induces the formation of protein bridges, the critical step of viral aggregation to facilitate its clearance. SP-D inhibits SARS-CoV-2 replication in Caco-2 cells (EC90 = 3.7 μg/mL). Therefore, SP-D recognizes and binds to the spike-(S)-protein of SARS-CoV-2 in vitro, initiates the aggregation, and inhibits viral replication in cells. Combined with the low levels of SP-D observed in COVID-19 patients, these results suggest that SP-D is important in the immune response to SARS-CoV-2 and that rhSP-D supplementation has the potential to be a novel class of anti-viral that will target SARS-CoV-2 infection.
Keywords: ACE2; COVID-19; SP-D; anti-inflammatory; spike-protein.
Conflict of interest statement
P.S.K. and R.A. are paid as part-time consultants for Airway Therapeutics Inc. which is developing SP-D as a human therapeutic agent. M.S. is the Chief Executive Officer at Airway Therapeutics Inc.
Figures

References
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

