MCN-CPI: Multiscale Convolutional Network for Compound-Protein Interaction Prediction
- PMID: 34439785
- PMCID: PMC8392217
- DOI: 10.3390/biom11081119
MCN-CPI: Multiscale Convolutional Network for Compound-Protein Interaction Prediction
Abstract
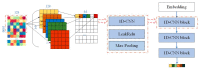
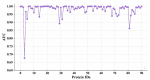
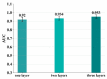
In the process of drug discovery, identifying the interaction between the protein and the novel compound plays an important role. With the development of technology, deep learning methods have shown excellent performance in various situations. However, the compound-protein interaction is complicated and the features extracted by most deep models are not comprehensive, which limits the performance to a certain extent. In this paper, we proposed a multiscale convolutional network that extracted the local and global features of the protein and the topological feature of the compound using different types of convolutional networks. The results showed that our model obtained the best performance compared with the existing deep learning methods.
Keywords: compound–protein interaction; convolutional network; deep learning; drug screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Wang S., Li Z., Zhang S., Jiang M., Wang X., Wei Z. Molecular Property Prediction Based on a Multichannel Substructure Graph. IEEE Access. 2020;8:18601–18614. doi: 10.1109/ACCESS.2020.2968535. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

