The Histone Variant MacroH2A1 Impacts Circadian Gene Expression and Cell Phenotype in an In Vitro Model of Hepatocellular Carcinoma
- PMID: 34440260
- PMCID: PMC8391426
- DOI: 10.3390/biomedicines9081057
The Histone Variant MacroH2A1 Impacts Circadian Gene Expression and Cell Phenotype in an In Vitro Model of Hepatocellular Carcinoma
Abstract
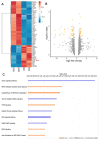
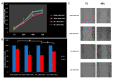
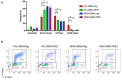
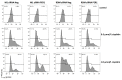
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related death worldwide. A foremost risk factor for HCC is obesity/metabolic syndrome-related non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH), which is prompted by remarkable changes in transcription patterns of genes enriching metabolic, immune/inflammatory, and circadian pathways. Epigenetic mechanisms play a role in NAFLD-associated HCC, and macroH2A1, a variant of histone H2A, is involved in the pathogenesis modulating the expression of oncogenes and/or tumor suppressor genes and interacting with SIRT1, which crucially impacts the circadian clock circuitry. Hence, we aimed to appraise if and how macroH2A1 regulated the expression patterns of circadian genes in the setting of NAFLD-associated HCC. We took advantage of an in vitro model of liver cancer represented by HepG2 (human hepatocarcinoma) cells stably knocked down for macroH2A1 and conducted whole transcriptome profiling and deep phenotyping analysis. We found up-regulation of PER1 along with several deregulated circadian genes, enriching several important pathways and functions related to cancer onset and progression, such as epithelial-to-mesenchymal transition, cell cycle deregulation, and DNA damage. PER1 silencing partially mitigated the malignant phenotype induced by the loss of macroH2A1 in HCC cells. In conclusion, our findings suggest a modulatory role for the core circadian protein PER1 in liver carcinogenesis in the context of a lack of the macroH2A1 epigenetic and transcriptional landscape.
Keywords: HCC; PER1; biological clock; cancer; circadian.
Conflict of interest statement
The authors declare that there are no conflict of interest with respect to the authorship and/or publication of this article.
Figures



References
-
- Akinyemiju T., Abera S., Ahmed M., Alam N., Alemayohu M.A., Allen C., Al-Raddadi R., Alvis-Guzman N., Amoako Y., Artaman A., et al. The Burden of Primary Liver Cancer and Underlying Etiologies from 1990 to 2015 at the Global, Regional, and National Level: Results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017;3:1683–1691. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

