Upregulation of 15 Antisense Long Non-Coding RNAs in Osteosarcoma
- PMID: 34440306
- PMCID: PMC8394133
- DOI: 10.3390/genes12081132
Upregulation of 15 Antisense Long Non-Coding RNAs in Osteosarcoma
Abstract
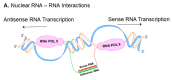
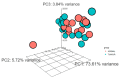
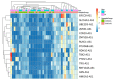
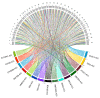
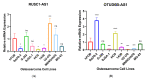
The human genome encodes thousands of natural antisense long noncoding RNAs (lncRNAs); they play the essential role in regulation of gene expression at multiple levels, including replication, transcription and translation. Dysregulation of antisense lncRNAs plays indispensable roles in numerous biological progress, such as tumour progression, metastasis and resistance to therapeutic agents. To date, there have been several studies analysing antisense lncRNAs expression profiles in cancer, but not enough to highlight the complexity of the disease. In this study, we investigated the expression patterns of antisense lncRNAs from osteosarcoma and healthy bone samples (24 tumour-16 bone samples) using RNA sequencing. We identified 15 antisense lncRNAs (RUSC1-AS1, TBX2-AS1, PTOV1-AS1, UBE2D3-AS1, ERCC8-AS1, ZMIZ1-AS1, RNF144A-AS1, RDH10-AS1, TRG-AS1, GSN-AS1, HMGA2-AS1, ZNF528-AS1, OTUD6B-AS1, COX10-AS1 and SLC16A1-AS1) that were upregulated in tumour samples compared to bone sample controls. Further, we performed real-time polymerase chain reaction (RT-qPCR) to validate the expressions of the antisense lncRNAs in 8 different osteosarcoma cell lines (SaOS-2, G-292, HOS, U2-OS, 143B, SJSA-1, MG-63, and MNNG/HOS) compared to hFOB (human osteoblast cell line). These differentially expressed IncRNAs can be considered biomarkers and potential therapeutic targets for osteosarcoma.
Keywords: alternative splicing; antisense RNA; non-coding RNA; osteosarcoma; sarcoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
A novel antisense long non-coding RNA SATB2-AS1 overexpresses in osteosarcoma and increases cell proliferation and growth.Mol Cell Biochem. 2017 Jun;430(1-2):47-56. doi: 10.1007/s11010-017-2953-9. Epub 2017 Feb 11. Mol Cell Biochem. 2017. PMID: 28190168
-
Upregulation of long non-coding RNA NNT-AS1 promotes osteosarcoma progression by inhibiting the tumor suppressive miR-320a.Cancer Biol Ther. 2019;20(4):413-422. doi: 10.1080/15384047.2018.1538612. Epub 2018 Nov 29. Cancer Biol Ther. 2019. PMID: 30489194 Free PMC article.
-
The role and clinical significance of long noncoding RNA zinc finger E-box-binding homeobox two antisense RNA 1 in promoting osteosarcoma cancer cell proliferation, inhibiting apoptosis and increasing migration by regulating miR-145.Anticancer Drugs. 2021 Feb 1;32(2):168-177. doi: 10.1097/CAD.0000000000000984. Anticancer Drugs. 2021. PMID: 32826416
-
The Emerging Role of Thymopoietin-Antisense RNA 1 as Long Noncoding RNA in the Pathogenesis of Human Cancers.DNA Cell Biol. 2021 Jul;40(7):848-857. doi: 10.1089/dna.2021.0024. Epub 2021 Jun 4. DNA Cell Biol. 2021. PMID: 34096793 Review.
-
The critical roles of lncRNAs in the development of osteosarcoma.Biomed Pharmacother. 2021 Mar;135:111217. doi: 10.1016/j.biopha.2021.111217. Epub 2021 Feb 1. Biomed Pharmacother. 2021. PMID: 33433358 Review.
Cited by
-
Unveiling the Genetic Complexity of Teratozoospermia: Integrated Genomic Analysis Reveals Novel Insights into lncRNAs' Role in Male Infertility.Int J Mol Sci. 2023 Oct 9;24(19):15002. doi: 10.3390/ijms241915002. Int J Mol Sci. 2023. PMID: 37834450 Free PMC article.
-
SEMA3G, downregulated by ncRNAs, correlates with favorable prognosis and tumor immune infiltration in kidney renal clear cell carcinoma.Aging (Albany NY). 2023 Dec 8;15(23):13944-13960. doi: 10.18632/aging.205277. Epub 2023 Dec 8. Aging (Albany NY). 2023. PMID: 38070142 Free PMC article.
-
Prognostic implications of necroptosis-related long noncoding RNA signatures in muscle-invasive bladder cancer.Front Genet. 2022 Dec 2;13:1036098. doi: 10.3389/fgene.2022.1036098. eCollection 2022. Front Genet. 2022. PMID: 36531246 Free PMC article.
-
The Role of Long Noncoding RNAs on Male Infertility: A Systematic Review and In Silico Analysis.Biology (Basel). 2022 Oct 15;11(10):1510. doi: 10.3390/biology11101510. Biology (Basel). 2022. PMID: 36290414 Free PMC article. Review.
-
Bioinformatics Analysis and Validation of the Role of Lnc-RAB11B-AS1 in the Development and Prognosis of Hepatocellular Carcinoma.Cells. 2022 Nov 6;11(21):3517. doi: 10.3390/cells11213517. Cells. 2022. PMID: 36359911 Free PMC article.
References
-
- Goryn T., Pienkowski A., Szostakowski B., Zdzienicki M., Lugowska I., Rutkowski P. Functional outcome of surgical treatment of adults with extremity osteosarcoma after megaprosthetic reconstruction-single-center experience. J. Orthop. Surg. Res. 2019;14:346. doi: 10.1186/s13018-019-1379-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

