Severity of Idiopathic Scoliosis Is Associated with Differential Methylation: An Epigenome-Wide Association Study of Monozygotic Twins with Idiopathic Scoliosis
- PMID: 34440365
- PMCID: PMC8391702
- DOI: 10.3390/genes12081191
Severity of Idiopathic Scoliosis Is Associated with Differential Methylation: An Epigenome-Wide Association Study of Monozygotic Twins with Idiopathic Scoliosis
Abstract
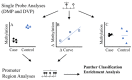
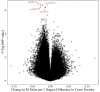
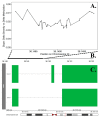
Epigenetic mechanisms may contribute to idiopathic scoliosis (IS). We identified 8 monozygotic twin pairs with IS, 6 discordant (Cobb angle difference > 10°) and 2 concordant (Cobb angle difference ≤ 2°). Genome-wide methylation in blood was measured with the Infinium HumanMethylation EPIC Beadchip. We tested for differences in methylation and methylation variability between discordant twins and tested the association between methylation and curve severity in all twins. Differentially methylated region (DMR) analyses identified gene promoter regions. Methylation at cg12959265 (chr. 7 DPY19L1) was less variable in cases (false discovery rate (FDR) = 0.0791). We identified four probes (false discovery rate, FDR < 0.10); cg02477677 (chr. 17, RARA gene), cg12922161 (chr. 2 LOC150622 gene), cg08826461 (chr. 2), and cg16382077 (chr. 7) associated with curve severity. We identified 57 DMRs where hyper- or hypo-methylation was consistent across the region and 28 DMRs with a consistent association with curve severity. Among DMRs, 21 were correlated with bone methylation. Prioritization of regions based on methylation concordance in bone identified promoter regions for WNT10A (WNT signaling), NPY (regulator of bone and energy homeostasis), and others predicted to be relevant for bone formation/remodeling. These regions may aid in understanding the complex interplay between genetics, environment, and IS.
Keywords: DNA methylation; bone; curve severity; differentially methylated region; discordant; epigenome-wide association study; idiopathic scoliosis; monozygotic twin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Khanshour A.M., Wise C.A. Pathogenesis of Idiopathic Scoliosis. Springer; Berlin, Germany: 2018. The Genetic Architecture of Adolescent Idiopathic Scoliosis; pp. 51–74.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

