DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting
- PMID: 34440388
- PMCID: PMC8394515
- DOI: 10.3390/genes12081214
DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting
Abstract
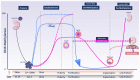
Genomic imprinting is an epigenetic marking process that results in the monoallelic expression of a subset of genes. Many of these 'imprinted' genes in mice and humans are involved in embryonic and extraembryonic growth and development, and some have life-long impacts on metabolism. During mammalian development, the genome undergoes waves of (re)programming of DNA methylation and other epigenetic marks. Disturbances in these events can cause imprinting disorders and compromise development. Multi-locus imprinting disturbance (MLID) is a condition by which imprinting defects touch more than one locus. Although most cases with MLID present with clinical features characteristic of one imprinting disorder. Imprinting defects also occur in 'molar' pregnancies-which are characterized by highly compromised embryonic development-and in other forms of reproductive compromise presenting clinically as infertility or early pregnancy loss. Pathogenic variants in some of the genes encoding proteins of the subcortical maternal complex (SCMC), a multi-protein complex in the mammalian oocyte, are responsible for a rare subgroup of moles, biparental complete hydatidiform mole (BiCHM), and other adverse reproductive outcomes which have been associated with altered imprinting status of the oocyte, embryo and/or placenta. The finding that defects in a cytoplasmic protein complex could have severe impacts on genomic methylation at critical times in gamete or early embryo development has wider implications beyond these relatively rare disorders. It signifies a potential for adverse maternal physiology, nutrition, or assisted reproduction to cause epigenetic defects at imprinted or other genes. Here, we review key milestones in DNA methylation patterning in the female germline and the embryo focusing on humans. We provide an overview of recent findings regarding DNA methylation deficits causing BiCHM, MLID, and early embryonic arrest. We also summarize identified SCMC mutations with regard to early embryonic arrest, BiCHM, and MLID.
Keywords: DNA methylation; embryo arrest; epigenetics; epimutations; genomic imprinting; infertility; oocyte; subcortical maternal complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

