Forecasting COVID-19 Severity by Intelligent Optical Fingerprinting of Blood Samples
- PMID: 34441244
- PMCID: PMC8392709
- DOI: 10.3390/diagnostics11081309
Forecasting COVID-19 Severity by Intelligent Optical Fingerprinting of Blood Samples
Abstract
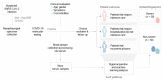
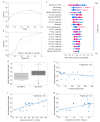
Forecasting COVID-19 disease severity is key to supporting clinical decision making and assisting resource allocation, particularly in intensive care units (ICUs). Here, we investigated the utility of time- and frequency-related features of the backscattered signal of serum patient samples to predict COVID-19 disease severity immediately after diagnosis. ICU admission was the primary outcome used to define disease severity. We developed a stacking ensemble machine learning model including the backscattered signal features (optical fingerprint), patient comorbidities, and age (AUROC = 0.80), which significantly outperformed the predictive value of clinical and laboratory variables available at hospital admission (AUROC = 0.71). The information derived from patient optical fingerprints was not strongly correlated with any clinical/laboratory variable, suggesting that optical fingerprinting brings unique information for COVID-19 severity risk assessment. Optical fingerprinting is a label-free, real-time, and low-cost technology that can be easily integrated as a front-line tool to facilitate the triage and clinical management of COVID-19 patients.
Keywords: COVID-19; machine learning; optical fingerprinting; photonics; predictive biomarker.
Conflict of interest statement
S.P.F., C.C., V.P., S.M.R., J.A., F.M., P.H.S., S.R., P.S., M.M and J.S.P. are employees and/or founders of iLoF. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., The Northwell COVID-19 Research Consortium Presenting characteristics, comorbidities and outcomes among 5700 patients hospitalized With COVID-19 in the New York City area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. - DOI - PMC - PubMed
-
- Helms J., Tacquard C., Severac F., Leonard-Lorant I., Ohana M., Delabranche X., Merdji H., Clere-Jehl R., Schenck M., Gandet F.F., et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: A multicenter prospective cohort study. Intensive Care Med. 2020;46:1089–1098. doi: 10.1007/s00134-020-06062-x. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

