Identification and Characterization of MicroRNAs in Gonads of Helicoverpa armigera (Lepidoptera: Noctuidae)
- PMID: 34442315
- PMCID: PMC8396854
- DOI: 10.3390/insects12080749
Identification and Characterization of MicroRNAs in Gonads of Helicoverpa armigera (Lepidoptera: Noctuidae)
Abstract
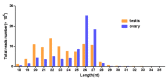
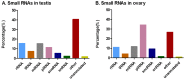
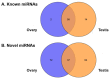
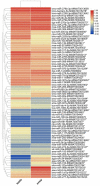
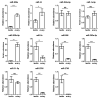
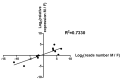
The high fecundity of the most destructive pest Helicoverpa armigera and its great resistance risk to insecticides and Bt crops make the reproductive-destruction-based control of this pest extremely appealing. To find suitable targets for disruption of its reproduction, we observed the testis and ovary development of H. armigera and conducted deep sequencing of the ovary and testis small RNAs of H. armigera and quantitative RT-PCR (RT-qPCR) validation to identify reproduction-related micro RNAs (miRNAs). A total of 7,592,150 and 8,815,237 clean reads were obtained from the testis and ovary tissue, respectively. After further analysis, we obtained 173 novel and 74 known miRNAs from the two libraries. Among the 74 known miRNAs, 60 miRNAs existed in the ovary and 72 existed in the testis. Further RT-qPCR validation of 5 miRNAs from the ovary and 6 miRNAs from the testis confirmed 8 of them were indeed ovary- (miR-989a, miR-263-5p, miR-34) or testis-biased (miR-2763, miR-998, miR-2c, miR-2765, miR-252a-5p). The 8 ovary- or testis-biased miRNAs had a total of 30,172 putative non-redundant target transcripts, as predicted by miRanda and RNAhybrid. Many of these target transcripts are assigned to reproduction-related GO terms (e.g., oocyte maturation, vitellogenesis, spermatogenesis) and are members of multiple reproduction-related KEGG pathways, such as the JAK-STAT signaling pathway, oocyte meiosis, the insulin signaling pathway, and insect hormone biosynthesis. These results suggest that the 8 gonad-biased miRNAs play important roles in reproduction and may be used as the targets for the development of reproductive-destruction-based control of H. armigera and, possibly, other lepidopteran pests.
Keywords: development; miRNAs; ovary; piRNAs; reproductive destruction; sex; testis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hombach S., Kretz M. Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 2016;937:3–17. - PubMed
-
- Yang J.X., Rastetter R.H., Wilhelm D. Non-coding RNAs: An introduction. Adv. Exp. Med. Biol. 2016;886:13–32. - PubMed
-
- Yuan Z.H., Zhao Y.M. The regulatory functions of piRNA/PIWI in spermatogenesis. Yi Chuan. 2017;39:683–691. - PubMed
Grants and funding
- hatch grant ARZT-1360890-H31-164 and multi state grant ARZT-1370400-R31-172 (NC246)/the USDA National Institute of Food and Agriculture
- No.U200420044/National Science Foundation of China (NSFC)-Henan Joint major grant
- No.21A210027/Key Scientific Research Projects of Colleges and Universities in Henan Province
- No.CB2020A06/State Key Laboratory of Cotton Biology Open Fund
LinkOut - more resources
Full Text Sources

