Deciphering Bacterial Community Structure, Functional Prediction and Food Safety Assessment in Fermented Fruits Using Next-Generation 16S rRNA Amplicon Sequencing
- PMID: 34442653
- PMCID: PMC8401261
- DOI: 10.3390/microorganisms9081574
Deciphering Bacterial Community Structure, Functional Prediction and Food Safety Assessment in Fermented Fruits Using Next-Generation 16S rRNA Amplicon Sequencing
Abstract
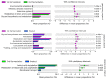
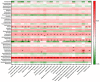
Fermented fruits and vegetables play an important role in safeguarding food security world-wide. Recently, robust sequencing-based microbial community analysis platforms have improved microbial safety assessment. This study aimed to examine the composition of bacteria and evaluate the bacterial safety of fermented fruit products using high-throughput 16S-rRNA metagenomic analysis. The operational taxonomic unit-based taxonomic classification of DNA sequences revealed 53 bacterial genera. However, the amplicon sequencing variant (ASV)-based clustering revealed 43 classifiable bacterial genera. Taxonomic classifications revealed that the abundance of Sphingomonas, which was the predominant genus in the majority of tested samples, was more than 85-90% among the total identified bacterial community in most samples. Among these identified genera, 13 low abundance genera were potential opportunistic pathogens, including Acinetobacter, Bacillus, Staphylococcus, Clostridium, Klebsiella, Mycobacterium, Ochrobactrum, Chryseobacterium, Stenotrophomonas, and Streptococcus. Of these 13 genera, 13 major opportunistic pathogenic species were validated using polymerase chain reaction. The pathogens were not detected in the samples of different stages and the final products of fermentation, except in one sample from the first stage of fermentation in which S. aureus was detected. This finding was consistent with that of ASV-based taxonomic classification according to which S. aureus was detected only in the sample from the first stage of fermentation. However, S. aureus was not significantly correlated with the human disease pathways. These results indicated that fermentation is a reliable and safe process as pathogenic bacteria were not detected in the fermentation products. The hybrid method reported in this study can be used simultaneously to evaluate the bacterial diversity, their functional predictions and safety assessment of novel fermentation products. Additionally, this hybrid method does not involve the random detection of pathogens, which can markedly decrease the time of detection and food safety verification. Furthermore, this hybrid method can be used for the quality control of products and the identification of external contamination.
Keywords: 16S rRNA metagenomics; bacterial diversity; fermented fruits; food safety; functional prediction; next-generation sequencing; opportunistic pathogens.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Battcock M. Fermented Fruits and Vegetables: A Global Perspective. Food & Agriculture Organization; Rome, Italy: 1998.
-
- Hampton J., Tang C., Jayasree Subhash A., Serventi L. Assessment of pear juice and puree as a fermentation matrix for water kefir. J. Food Process. Preserv. 2021;45:e15223. doi: 10.1111/jfpp.15223. - DOI
-
- Shah N.N., Singhal R.S. 3—Fermented Fruits and Vegetables. In: Pandey A., Sanromán M.Á., Du G., Soccol C.R., Dussap C.-G., editors. Current Developments in Biotechnology and Bioengineering. Elsevier; Amsterdam, The Netherlands: 2017. pp. 45–89.
LinkOut - more resources
Full Text Sources

