The Sisal Virome: Uncovering the Viral Diversity of Agave Varieties Reveals New and Organ-Specific Viruses
- PMID: 34442783
- PMCID: PMC8400513
- DOI: 10.3390/microorganisms9081704
The Sisal Virome: Uncovering the Viral Diversity of Agave Varieties Reveals New and Organ-Specific Viruses
Abstract
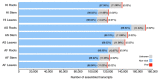
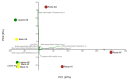
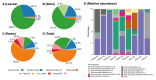
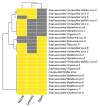
Sisal is a common name for different plant varieties in the genus Agave (especially Agave sisalana) used for high-quality natural leaf fiber extraction. Despite the economic value of these plants, we still lack information about the diversity of viruses (virome) in non-tequilana species from the genus Agave. In this work, by associating RNA and DNA deep sequencing we were able to identify 25 putative viral species infecting A. sisalana, A. fourcroydes, and Agave hybrid 11648, including one strain of Cowpea Mild Mottle Virus (CPMMV) and 24 elements likely representing new viruses. Phylogenetic analysis indicated they belong to at least six viral families: Alphaflexiviridae, Betaflexiviridae, Botourmiaviridae, Closteroviridae, Partitiviridae, Virgaviridae, and three distinct unclassified groups. We observed higher viral taxa richness in roots when compared to leaves and stems. Furthermore, leaves and stems are very similar diversity-wise, with a lower number of taxa and dominance of a single viral species. Finally, approximately 50% of the identified viruses were found in all Agave organs investigated, which suggests that they likely produce a systemic infection. This is the first metatranscriptomics study focused on viral identification in species from the genus Agave. Despite having analyzed symptomless individuals, we identified several viruses supposedly infecting Agave species, including organ-specific and systemic species. Surprisingly, some of these putative viruses are probably infecting microorganisms composing the plant microbiota. Altogether, our results reinforce the importance of unbiased strategies for the identification and monitoring of viruses in plant species, including those with asymptomatic phenotypes.
Keywords: Agave; metatranscriptomics; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A novel tepovirus, Agave virus T, identified by the analysis of the transcriptome data of blue agave (Agave tequilana).Acta Virol. 2021;65(1):68-71. doi: 10.4149/av_2021_107. Acta Virol. 2021. PMID: 33827223
-
Unmapped RNA Virus Diversity in Termites and their Symbionts.Viruses. 2020 Oct 9;12(10):1145. doi: 10.3390/v12101145. Viruses. 2020. PMID: 33050289 Free PMC article.
-
Transcriptome Comparison Reveals Distinct Selection Patterns in Domesticated and Wild Agave Species, the Important CAM Plants.Int J Genomics. 2018 Nov 22;2018:5716518. doi: 10.1155/2018/5716518. eCollection 2018. Int J Genomics. 2018. PMID: 30596084 Free PMC article.
-
Viruses infecting macrofungi.Virusdisease. 2018 Mar;29(1):1-18. doi: 10.1007/s13337-018-0434-8. Epub 2018 Feb 8. Virusdisease. 2018. PMID: 29607353 Free PMC article. Review.
-
Advances in the Micropropagation and Genetic Transformation of Agave Species.Plants (Basel). 2022 Jul 1;11(13):1757. doi: 10.3390/plants11131757. Plants (Basel). 2022. PMID: 35807709 Free PMC article. Review.
Cited by
-
Seven new mycoviruses identified from isolated ascomycetous macrofungi.Virus Res. 2024 Jan 2;339:199290. doi: 10.1016/j.virusres.2023.199290. Epub 2023 Dec 10. Virus Res. 2024. PMID: 38043725 Free PMC article.
-
Molecular characterization of a novel virga-like virus associated with wheat.Arch Virol. 2022 Sep;167(9):1909-1913. doi: 10.1007/s00705-022-05473-z. Epub 2022 Jun 25. Arch Virol. 2022. PMID: 35752685
-
A group of segmented viruses contains genome segments sharing homology with multiple viral taxa.J Virol. 2025 Jul 22;99(7):e0033225. doi: 10.1128/jvi.00332-25. Epub 2025 Jun 4. J Virol. 2025. PMID: 40464578 Free PMC article.
-
Deciphering the virome of Chunkung (Cnidium officinale) showing dwarfism-like symptoms via a high-throughput sequencing analysis.Virol J. 2024 Apr 15;21(1):86. doi: 10.1186/s12985-024-02361-7. Virol J. 2024. PMID: 38622686 Free PMC article.
-
High-Throughput Sequencing Detects a Viral Complex in Agave Tequilana Plants.Adv Virol. 2025 May 4;2025:6434701. doi: 10.1155/av/6434701. eCollection 2025. Adv Virol. 2025. PMID: 40351455 Free PMC article.
References
-
- Brown K. Agave sisalana Perrine. Wildl. Weeds. 2002;5:18–21.
-
- González-Iturbe J.A., Olmsted I., Tun-Dzul F. Tropical dry forest recovery after long term Henequen (sisal, Agave fourcroydes Lem.) plantation in northern Yucatan, Mexico. For. Ecol. Manag. 2002;167:67–82. doi: 10.1016/S0378-1127(01)00689-2. - DOI
-
- Santos E.M.C., Silva O.A. Da Sisal in Bahia—Brazil. Mercator. 2017;16:1–13. doi: 10.4215/rm2017.e16029. - DOI
-
- De Andrade Santos R., Brandão W. ÁRVORE DO CONHECIMENTO: Território Sisal. EMBRAPA. [(accessed on 31 May 2021)]; Available online: https://www.agencia.cnptia.embrapa.br/gestor/territorio_sisal/arvore/CON....
-
- Alves J.J.A., de Araújo M.A., do Nascimento S.S. Degradação da caatinga: Uma investigação ecogeográfica. Rev. Caatinga. 2009;22:126–135.
Grants and funding
LinkOut - more resources
Full Text Sources

