The Effect of the Anticipated Nuclear Localization Sequence of ' Candidatus Phytoplasma mali' SAP11-like Protein on Localization of the Protein and Destabilization of TCP Transcription Factor
- PMID: 34442835
- PMCID: PMC8401217
- DOI: 10.3390/microorganisms9081756
The Effect of the Anticipated Nuclear Localization Sequence of ' Candidatus Phytoplasma mali' SAP11-like Protein on Localization of the Protein and Destabilization of TCP Transcription Factor
Abstract
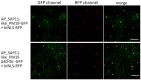
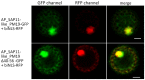
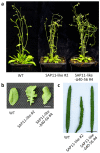
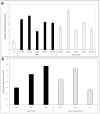
SAP11 is an effector protein that has been identified in various phytoplasma species. It localizes in the plant nucleus and can bind and destabilize TEOSINE BRANCHES/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) transcription factors. Although SAP11 of different phytoplasma species share similar activities, their protein sequences differ greatly. Here, we demonstrate that the SAP11-like protein of 'Candidatus Phytoplasma mali' ('Ca. P. mali') strain PM19 localizes into the plant nucleus without requiring the anticipated nuclear localization sequence (NLS). We show that the protein induces crinkled leaves and siliques, and witches' broom symptoms, in transgenic Arabidopsis thaliana (A. thaliana) plants and binds to six members of class I and all members of class II TCP transcription factors of A. thaliana in yeast two-hybrid assays. We also identified a 17 amino acid stretch previously predicted to be a nuclear localization sequence that is important for the binding of some of the TCPs, which results in a crinkled leaf and silique phenotype in transgenic A. thaliana. Moreover, we provide evidence that the SAP11-like protein has a destabilizing effect on some TCPs in vivo.
Keywords: SAP11; TCP transcription factors; phytoplasmas; plant-pathogen interaction; ‘Candidatus Phytoplasma mali’.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bai X., Zhang J., Ewing A., Miller S.A., Jancso Radek A., Shevchenko D.V., Tsukerman K., Walunas T., Lapidus A., Campbell J.W., et al. Living with genome instability: The adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 2006;188:3682–3696. doi: 10.1128/JB.188.10.3682-3696.2006. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

