Clavibacter michiganensis Downregulates Photosynthesis and Modifies Monolignols Metabolism Revealing a Crosstalk with Tomato Immune Responses
- PMID: 34445148
- PMCID: PMC8395114
- DOI: 10.3390/ijms22168442
Clavibacter michiganensis Downregulates Photosynthesis and Modifies Monolignols Metabolism Revealing a Crosstalk with Tomato Immune Responses
Abstract
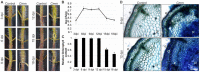
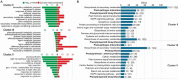
The gram-positive pathogenic bacterium Clavibacter michiganensis subsp. michiganensis (Cmm) causes bacterial canker disease in tomato, affecting crop yield and fruit quality. To understand how tomato plants respond, the dynamic expression profile of host genes was analyzed upon Cmm infection. Symptoms of bacterial canker became evident from the third day. As the disease progressed, the bacterial population increased in planta, reaching the highest level at six days and remained constant till the twelfth day post inoculation. These two time points were selected for transcriptomics. A progressive down-regulation of key genes encoding for components of the photosynthetic apparatus was observed. Two temporally separated defense responses were observed, which were to an extent interdependent. During the primary response, genes of the phenylpropanoid pathway were diverted towards the synthesis of monolignols away from S-lignin. In dicots, lignin polymers mainly consist of G- and S-units, playing an important role in defense. The twist towards G-lignin enrichment is consistent with previous findings, highlighting a response to generate an early protective barrier and to achieve a tight interplay between lignin recomposition and the primary defense response mechanism. Upon progression of Cmm infection, the temporal deactivation of phenylpropanoids coincided with the upregulation of genes that belong in a secondary response mechanism, supporting an elegant reprogramming of the host transcriptome to establish a robust defense apparatus and suppress pathogen invasion. This high-throughput analysis reveals a dynamic reorganization of plant defense mechanisms upon bacterial infection to implement an array of barriers preventing pathogen invasion and spread.
Keywords: RNA-seq; Solanum lycopersicum; defense lignin; gram-positive; monolignols; phenylpropanoids; photosynthesis; plant immunity; plant pathogen interaction; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Clavibacter michiganensis Reframed: The Story of How the Genomics Era Made a New Face for an Old Enemy.Mol Plant Pathol. 2025 May;26(5):e70093. doi: 10.1111/mpp.70093. Mol Plant Pathol. 2025. PMID: 40391582 Free PMC article. Review.
-
Transcriptome analysis of Clavibacter michiganensis subsp. michiganensis-infected tomatoes: a role of salicylic acid in the host response.BMC Plant Biol. 2021 Oct 19;21(1):476. doi: 10.1186/s12870-021-03251-8. BMC Plant Biol. 2021. PMID: 34666675 Free PMC article.
-
Comparative RNA-Seq Analysis Reveals Potentially Resistance-Related Genes in Response to Bacterial Canker of Tomato.Genes (Basel). 2021 Oct 29;12(11):1745. doi: 10.3390/genes12111745. Genes (Basel). 2021. PMID: 34828351 Free PMC article.
-
Comparative transcriptome analysis of resistant and cultivated tomato lines in response to Clavibacter michiganensis subsp. michiganensis.Genomics. 2021 Jul;113(4):2455-2467. doi: 10.1016/j.ygeno.2021.05.033. Epub 2021 May 28. Genomics. 2021. PMID: 34052318
-
Clavibacter michiganensis subsp. michiganensis: first steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium.J Biotechnol. 2003 Dec 19;106(2-3):179-91. doi: 10.1016/j.jbiotec.2003.07.011. J Biotechnol. 2003. PMID: 14651860 Review.
Cited by
-
Transcriptome analysis of tomato plants following salicylic acid-induced immunity against Clavibacter michiganensis ssp. michiganensis.Plant Biotechnol (Tokyo). 2023 Dec 25;40(4):273-282. doi: 10.5511/plantbiotechnology.23.0711a. Plant Biotechnol (Tokyo). 2023. PMID: 38434116 Free PMC article.
-
CRISPR/dCas12a-mediated activation of SlPAL2 enhances tomato resistance against bacterial canker disease.PLoS One. 2025 Mar 26;20(3):e0320436. doi: 10.1371/journal.pone.0320436. eCollection 2025. PLoS One. 2025. PMID: 40138366 Free PMC article.
-
Transcriptomics combined with physiological analysis provides insights into the mechanism of resistance to Coleosporium bletiae in Bletilla striata.Front Plant Sci. 2025 Jul 14;16:1604512. doi: 10.3389/fpls.2025.1604512. eCollection 2025. Front Plant Sci. 2025. PMID: 40726552 Free PMC article.
-
Clavibacter michiganensis Reframed: The Story of How the Genomics Era Made a New Face for an Old Enemy.Mol Plant Pathol. 2025 May;26(5):e70093. doi: 10.1111/mpp.70093. Mol Plant Pathol. 2025. PMID: 40391582 Free PMC article. Review.
References
-
- Martínez-Valverde I., Periago M.J., Provan G., Chesson A. Phenolic compounds, lycopene and antioxidant activity in commercial varieties of tomato (Lycopersicum esculentum) J. Sci. Food Agric. 2002;82:323–330. doi: 10.1002/jsfa.1035. - DOI
-
- Goode M.J., Sasser M. Prevention-the key to controlling bacterial spot and bacterial speck of tomato. Plant Dis. 1980;64:831–834. doi: 10.1094/PD-64-831. - DOI
-
- Jones J.B., Zitter T.A., Momol T.M., Miller S.A. Compendium of Tomato Diseases and Pests. 2nd ed. APS Press; Eagan, MN, USA: 2014.
-
- Davis M.J., Gillaspie A.G., Vidaner A.K., Jr., Harris R.W. Clavibacter: A new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int. J. Syst. Bacteriol. 1984;34:107–111.
-
- Smith E.F. A new tomato disease of economic importance. Science. 1910;31:794–796.
MeSH terms
Supplementary concepts
Grants and funding
- T1EDK-04142/The European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH-CREATE-INNOVATE
- MIS: 5048459/Greece and the European Union (European Social Fund) through the Operational Program «Human Resources Development, Education and Lifelong Learning 2014-2020» and the Program encoded EDBM103, entitled "Support for researchers with an emphasis on young rese
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

