YY1 Silencing Induces 5-Fluorouracil-Resistance and BCL2L15 Downregulation in Colorectal Cancer Cells: Diagnostic and Prognostic Relevance
- PMID: 34445183
- PMCID: PMC8395225
- DOI: 10.3390/ijms22168481
YY1 Silencing Induces 5-Fluorouracil-Resistance and BCL2L15 Downregulation in Colorectal Cancer Cells: Diagnostic and Prognostic Relevance
Abstract
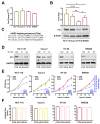
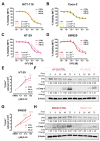
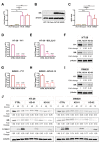
Colorectal cancer (CRC) is characterized by genetic heterogeneity and is often diagnosed at an advanced stage. Therefore, there is a need to identify novel predictive markers. Yin Yang 1 (YY1) is a transcription factor playing a dual role in cancer. The present study aimed to investigate whether YY1 expression levels influence CRC cell response to therapy and to identify the transcriptional targets involved. The diagnostic and prognostic values of YY1 and the identified factor(s) in CRC patients were also explored. Silencing of YY1 increased the resistance to 5-Fluorouracil-induced cytotoxicity in two out of four CRC cells with different genotypes. BCL2L15/Bfk pro-apoptotic factor was found selectively expressed in the responder CRC cells and downregulated upon YY1 knockdown. CRC dataset analyses corroborated a tumor-suppressive role for both YY1 and BCL2L15 whose expressions were inversely correlated with aggressiveness. CRC single-cell sequencing dataset analyses demonstrated higher co-expression levels of both YY1 and BCL2L15 within defined tumor cell clusters. Finally, elevated levels of YY1 and BCL2L15 in CRC patients were associated with larger relapse-free survival. Given their observed anti-cancer role, we propose YY1 and BCL2L15 as candidate diagnostic and prognostic CRC biomarkers.
Keywords: BCL2L15/Bfk; Yin Yang 1; biomarkers; colorectal cancer; tumor-suppressor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

