The Contribution of Autophagy and LncRNAs to MYC-Driven Gene Regulatory Networks in Cancers
- PMID: 34445233
- PMCID: PMC8395220
- DOI: 10.3390/ijms22168527
The Contribution of Autophagy and LncRNAs to MYC-Driven Gene Regulatory Networks in Cancers
Abstract
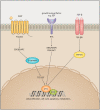
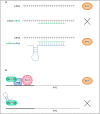
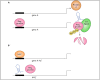
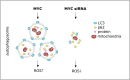
MYC is a target of the Wnt signalling pathway and governs numerous cellular and developmental programmes hijacked in cancers. The amplification of MYC is a frequently occurring genetic alteration in cancer genomes, and this transcription factor is implicated in metabolic reprogramming, cell death, and angiogenesis in cancers. In this review, we analyse MYC gene networks in solid cancers. We investigate the interaction of MYC with long non-coding RNAs (lncRNAs). Furthermore, we investigate the role of MYC regulatory networks in inducing changes to cellular processes, including autophagy and mitophagy. Finally, we review the interaction and mutual regulation between MYC and lncRNAs, and autophagic processes and analyse these networks as unexplored areas of targeting and manipulation for therapeutic gain in MYC-driven malignancies.
Keywords: MYC; autophagy; gene regulatory networks (GRNs); lncRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

