Hypomethylated RRBP1 Potentiates Tumor Malignancy and Chemoresistance in Upper Tract Urothelial Carcinoma
- PMID: 34445467
- PMCID: PMC8395942
- DOI: 10.3390/ijms22168761
Hypomethylated RRBP1 Potentiates Tumor Malignancy and Chemoresistance in Upper Tract Urothelial Carcinoma
Abstract
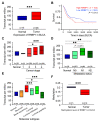
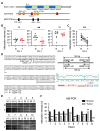
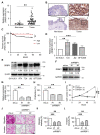
Ribosome-binding protein 1 (RRBP1) is a potential oncogene in several cancer types. However, the correlation between RRBP1 expression and the prognosis of patients with upper tract urothelial carcinoma (UTUC) remains unclear. In this study, we identified that RRBP1 is associated with carcinogenesis and metastasis in UTUC using a methylation profiling microarray. High correlations between RRBP1 and cancer stages, nodal metastasis status, molecular subtypes, and prognosis in bladder urothelial cancer (BLCA) were found. Aberrant DNA methylation in the gene body region of RRBP1 was determined in UTUC tissues by methylation-specific PCR. RRBP1 expression was significantly increased in UTUC tissues and cell lines, as determined by real-time PCR and immunohistochemistry. RRBP1 depletion significantly reduced BFTC909 cell growth induced by specific shRNA. On the other hand, molecular subtype analysis showed that the expression of RRBP1 was associated with genes related to cell proliferation, epithelial-mesenchymal transition, and basal markers. A patient-derived organoid model was established to analyze patients' responses to different drugs. The expression of RRBP1 was related to chemoresistance. Taken together, these results provide the first evidence that RRBP1 gene body hypomethylation predicts RRBP1 high expression in UTUC. The data highlight the importance of RRBP1 in UTUC malignancy and chemotherapeutic tolerance.
Keywords: RRBP1; chemoresistance; hypomethylation; oncogene; patient-derived organoid; upper tract urothelial carcinoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Rouprêt M., Babjuk M., Compérat E., Zigeuner R., Sylvester R.J., Burger M., Cowan N.C., Böhle A., van Rhijn B.W., Kaasinen E., et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Cell Carcinoma: 2015 Update. Eur. Urol. 2015;68:868–879. doi: 10.1016/j.eururo.2015.06.044. - DOI - PubMed
-
- Cha E.K., Shariat S.F., Kormaksson M., Novara G., Rink M., Lotan Y., Scherr D., Raman J.D., Kassouf W., Zigeuner R., et al. Predicting clinical outcomes after radical nephroureterectomy for upper tract urothelial carcinoma. J. Clin. Oncol. 2012;30:267. doi: 10.1200/jco.2012.30.5_suppl.267. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

