TRIM25 and DEAD-Box RNA Helicase DDX3X Cooperate to Regulate RIG-I-Mediated Antiviral Immunity
- PMID: 34445801
- PMCID: PMC8396550
- DOI: 10.3390/ijms22169094
TRIM25 and DEAD-Box RNA Helicase DDX3X Cooperate to Regulate RIG-I-Mediated Antiviral Immunity
Abstract
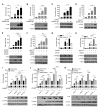
The cytoplasmic retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs) initiate interferon (IFN) production and antiviral gene expression in response to RNA virus infection. Consequently, RLR signalling is tightly regulated by both host and viral factors. Tripartite motif protein 25 (TRIM25) is an E3 ligase that ubiquitinates multiple substrates within the RLR signalling cascade, playing both ubiquitination-dependent and -independent roles in RIG-I-mediated IFN induction. However, additional regulatory roles are emerging. Here, we show a novel interaction between TRIM25 and another protein in the RLR pathway that is essential for type I IFN induction, DEAD-box helicase 3X (DDX3X). In vitro assays and knockdown studies reveal that TRIM25 ubiquitinates DDX3X at lysine 55 (K55) and that TRIM25 and DDX3X cooperatively enhance IFNB1 induction following RIG-I activation, but the latter is independent of TRIM25's catalytic activity. Furthermore, we found that the influenza A virus non-structural protein 1 (NS1) disrupts the TRIM25:DDX3X interaction, abrogating both TRIM25-mediated ubiquitination of DDX3X and cooperative activation of the IFNB1 promoter. Thus, our results reveal a new interplay between two RLR-host proteins that cooperatively enhance IFN-β production. We also uncover a new and further mechanism by which influenza A virus NS1 suppresses host antiviral defence.
Keywords: DDX3X; DEAD-box helicase; E3 ligase; IFN; NS1; RLR signalling; TRIM25; antiviral immunity; influenza; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Van Zuylen W.J., Doyon P., Clément J.-F., Khan K.A., D’Ambrosio L.M., Dô F., St-Amant-Verret M., Wissanji T., Emery G., Gingras A.-C., et al. Proteomic profiling of the TRAF3 interactome network reveals a new role for the ER-to-Golgi transport compartments in innate immunity. PLoS Pathog. 2012;8:e1002747. doi: 10.1371/journal.ppat.1002747. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

