Unexpected organellar locations of ESCRT machinery in Giardia intestinalis and complex evolutionary dynamics spanning the transition to parasitism in the lineage Fornicata
- PMID: 34446013
- PMCID: PMC8394649
- DOI: 10.1186/s12915-021-01077-2
Unexpected organellar locations of ESCRT machinery in Giardia intestinalis and complex evolutionary dynamics spanning the transition to parasitism in the lineage Fornicata
Abstract
Background: Comparing a parasitic lineage to its free-living relatives is a powerful way to understand how that evolutionary transition to parasitism occurred. Giardia intestinalis (Fornicata) is a leading cause of gastrointestinal disease world-wide and is famous for its unusual complement of cellular compartments, such as having peripheral vacuoles instead of typical endosomal compartments. Endocytosis plays an important role in Giardia's pathogenesis. Endosomal sorting complexes required for transport (ESCRT) are membrane-deforming proteins associated with the late endosome/multivesicular body (MVB). MVBs are ill-defined in G. intestinalis, and roles for identified ESCRT-related proteins are not fully understood in the context of its unique endocytic system. Furthermore, components thought to be required for full ESCRT functionality have not yet been documented in this species.
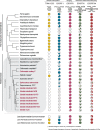
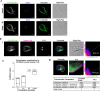
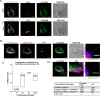
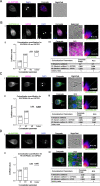
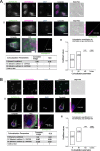
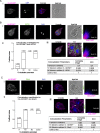
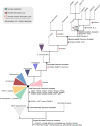
Results: We used genomic and transcriptomic data from several Fornicata species to clarify the evolutionary genome streamlining observed in Giardia, as well as to detect any divergent orthologs of the Fornicata ESCRT subunits. We observed differences in the ESCRT machinery complement between Giardia strains. Microscopy-based investigations of key components of ESCRT machinery such as GiVPS36 and GiVPS25 link them to peripheral vacuoles, highlighting these organelles as simplified MVB equivalents. Unexpectedly, we show ESCRT components associated with the endoplasmic reticulum and, for the first time, mitosomes. Finally, we identified the rare ESCRT component CHMP7 in several fornicate representatives, including Giardia and show that contrary to current understanding, CHMP7 evolved from a gene fusion of VPS25 and SNF7 domains, prior to the last eukaryotic common ancestor, over 1.5 billion years ago.
Conclusions: Our findings show that ESCRT machinery in G. intestinalis is far more varied and complete than previously thought, associates to multiple cellular locations, and presents changes in ESCRT complement which pre-date adoption of a parasitic lifestyle.
Keywords: ESCRT; Endomembrane; Evolutionary Cell Biology; Excavata; Giardia; PV; Parasitism.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
The minimal ESCRT machinery of Giardia lamblia has altered inter-subunit interactions within the ESCRT-II and ESCRT-III complexes.Eur J Cell Biol. 2018 Jan;97(1):44-62. doi: 10.1016/j.ejcb.2017.11.004. Epub 2017 Dec 6. Eur J Cell Biol. 2018. PMID: 29224850
-
The reduced ARF regulatory system in Giardia intestinalis pre-dates the transition to parasitism in the lineage Fornicata.Int J Parasitol. 2021 Sep;51(10):825-839. doi: 10.1016/j.ijpara.2021.02.004. Epub 2021 Apr 20. Int J Parasitol. 2021. PMID: 33848497
-
Significantly Diverged Did2/Vps46 Orthologues from the Protozoan Parasite Giardia lamblia.Curr Microbiol. 2015 Sep;71(3):333-40. doi: 10.1007/s00284-015-0844-4. Epub 2015 Jun 12. Curr Microbiol. 2015. PMID: 26068593
-
Molecular interplays of the Entamoeba histolytica endosomal sorting complexes required for transport during phagocytosis.Front Cell Infect Microbiol. 2022 Oct 27;12:855797. doi: 10.3389/fcimb.2022.855797. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36389174 Free PMC article. Review.
-
A cytonaut's guide to protein trafficking in Giardia lamblia.Adv Parasitol. 2019;106:105-127. doi: 10.1016/bs.apar.2019.08.001. Adv Parasitol. 2019. PMID: 31630756 Review.
Cited by
-
A core UPS molecular complement implicates unique endocytic compartments at the parasite-host interface in Giardia lamblia.Virulence. 2023 Dec;14(1):2174288. doi: 10.1080/21505594.2023.2174288. Virulence. 2023. PMID: 36730629 Free PMC article.
-
Evolutionary diversification of the autophagy-related ubiquitin-like conjugation systems.Autophagy. 2022 Dec;18(12):2969-2984. doi: 10.1080/15548627.2022.2059168. Epub 2022 Apr 15. Autophagy. 2022. PMID: 35427200 Free PMC article.
-
Genomic survey maps differences in the molecular complement of vesicle formation machinery between Giardia intestinalis assemblages.PLoS Negl Trop Dis. 2023 Dec 18;17(12):e0011837. doi: 10.1371/journal.pntd.0011837. eCollection 2023 Dec. PLoS Negl Trop Dis. 2023. PMID: 38109380 Free PMC article.
-
The retromer and retriever systems are conserved and differentially expanded in parabasalids.J Cell Sci. 2024 Jul 1;137(13):jcs261949. doi: 10.1242/jcs.261949. Epub 2024 Jul 12. J Cell Sci. 2024. PMID: 38884339 Free PMC article.
-
Combined nanometric and phylogenetic analysis of unique endocytic compartments in Giardia lamblia sheds light on the evolution of endocytosis in Metamonada.BMC Biol. 2022 Sep 21;20(1):206. doi: 10.1186/s12915-022-01402-3. BMC Biol. 2022. PMID: 36127707 Free PMC article.
References
-
- Allain T, Buret AG. Chapter Five - Pathogenesis and post-infectious complications in giardiasis. In: Ortega-Pierres MG, editor. Advances in Parasitology. Academic Press; 2020. pp. 173–199. - PubMed
-
- Faso C, Hehl AB. Advances in parasitology. Elsevier; 2019. A cytonaut’s guide to protein trafficking in Giardia lamblia; pp. 105–127. - PubMed
-
- Gargantini PR, Serradell M Del C, Ríos DN, Tenaglia AH, Luján HD. Antigenic variation in the intestinal parasite Giardia lamblia. Host-Microbe Interact Parasitesfungiviruses. 2016;32:52–58. - PubMed

