Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy
- PMID: 34446607
- PMCID: PMC9503018
- DOI: 10.1126/science.abc9113
Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy
Abstract
The antitumor efficacy of cancer immunotherapy can correlate with the presence of certain bacterial species within the gut microbiome. However, many of the molecular mechanisms that influence host response to immunotherapy remain elusive. In this study, we show that members of the bacterial genus Enterococcus improve checkpoint inhibitor immunotherapy in mouse tumor models. Active enterococci express and secrete orthologs of the NlpC/p60 peptidoglycan hydrolase SagA that generate immune-active muropeptides. Expression of SagA in nonprotective E. faecalis was sufficient to promote immunotherapy response, and its activity required the peptidoglycan sensor NOD2. Notably, SagA-engineered probiotics or synthetic muropeptides also augmented anti-PD-L1 antitumor efficacy. Taken together, our data suggest that microbiota species with specialized peptidoglycan remodeling activity and muropeptide-based therapeutics may enhance cancer immunotherapy and could be leveraged as next-generation adjuvants.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
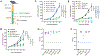
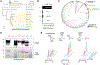


References
-
- Sahin U, Türeci Ö, Personalized vaccines for cancer immunotherapy. Science. 359, 1355–1360 (2018). - PubMed
-
- June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC, CAR T cell immunotherapy for human cancer. Science. 359, 1361–1365 (2018). - PubMed
-
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA, Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 350, 207–211 (2015). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

