The microbiome extends host evolutionary potential
- PMID: 34446709
- PMCID: PMC8390463
- DOI: 10.1038/s41467-021-25315-x
The microbiome extends host evolutionary potential
Abstract
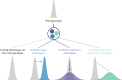
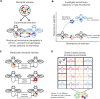
The microbiome shapes many host traits, yet the biology of microbiomes challenges traditional evolutionary models. Here, we illustrate how integrating the microbiome into quantitative genetics can help untangle complexities of host-microbiome evolution. We describe two general ways in which the microbiome may affect host evolutionary potential: by shifting the mean host phenotype and by changing the variance in host phenotype in the population. We synthesize the literature across diverse taxa and discuss how these scenarios could shape the host response to selection. We conclude by outlining key avenues of research to improve our understanding of the complex interplay between hosts and microbiomes.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Friesen ML, et al. Microbially mediated plant functional traits. Annu. Rev. Ecol. Evol. Syst. 2011;42:23–46. doi: 10.1146/annurev-ecolsys-102710-145039. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

