AHR signaling is induced by infection with coronaviruses
- PMID: 34446714
- PMCID: PMC8390748
- DOI: 10.1038/s41467-021-25412-x
AHR signaling is induced by infection with coronaviruses
Abstract
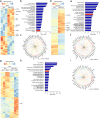
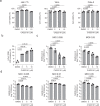
Coronavirus infection in humans is usually associated to respiratory tract illnesses, ranging in severity from mild to life-threatening respiratory failure. The aryl hydrocarbon receptor (AHR) was recently identified as a host factor for Zika and dengue viruses; AHR antagonists boost antiviral immunity, decrease viral titers and ameliorate Zika-induced pathology in vivo. Here we report that AHR is activated by infection with different coronaviruses, potentially impacting antiviral immunity and lung epithelial cells. Indeed, the analysis of single-cell RNA-seq from lung tissue detected increased expression of AHR and AHR transcriptional targets, suggesting AHR signaling activation in SARS-CoV-2-infected epithelial cells from COVID-19 patients. Moreover, we detected an association between AHR expression and viral load in SARS-CoV-2 infected patients. Finally, we found that the pharmacological inhibition of AHR suppressed the replication in vitro of one of the causative agents of the common cold, HCoV-229E, and the causative agent of the COVID-19 pandemic, SARS-CoV-2. Taken together, these findings suggest that AHR activation is a common strategy used by coronaviruses to evade antiviral immunity and promote viral replication, which may also contribute to lung pathology. Future studies should further evaluate the potential of AHR as a target for host-directed antiviral therapy.
© 2021. The Author(s).
Conflict of interest statement
F.J.Q. is a member of the Scientific Advisory Board of Kyn Therapeutics. The other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

