Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction
- PMID: 34446935
- PMCID: PMC8484054
- DOI: 10.1038/s41593-021-00908-3
Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction
Erratum in
-
Author Correction: Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction.Nat Neurosci. 2025 Jan;28(1):213. doi: 10.1038/s41593-024-01853-7. Nat Neurosci. 2025. PMID: 39643700 No abstract available.
Abstract
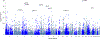
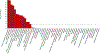
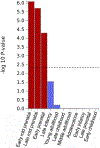
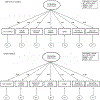
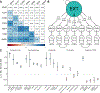
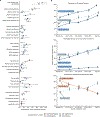
Behaviors and disorders related to self-regulation, such as substance use, antisocial behavior and attention-deficit/hyperactivity disorder, are collectively referred to as externalizing and have shared genetic liability. We applied a multivariate approach that leverages genetic correlations among externalizing traits for genome-wide association analyses. By pooling data from ~1.5 million people, our approach is statistically more powerful than single-trait analyses and identifies more than 500 genetic loci. The loci were enriched for genes expressed in the brain and related to nervous system development. A polygenic score constructed from our results predicts a range of behavioral and medical outcomes that were not part of genome-wide analyses, including traits that until now lacked well-performing polygenic scores, such as opioid use disorder, suicide, HIV infections, criminal convictions and unemployment. Our findings are consistent with the idea that persistent difficulties in self-regulation can be conceptualized as a neurodevelopmental trait with complex and far-reaching social and health correlates.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Figures

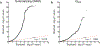
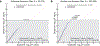



References
-
- Achenbach TM The classification of children’s psychiatric symptoms: A factor-analytic study. Psychol. Monogr. Gen. Appl 80, 1–37 (1966). - PubMed
-
- Hicks BM, Krueger RF, Iacono WG, McGue M & Patrick CJ Family transmission and heritability of externalizing disorders: a twin-family study. Arch. Gen. Psychiatry 61, 922–928 (2004). - PubMed
-
- Krueger RF et al.Etiologic connections among substance dependence, antisocial behavior and personality: Modeling the externalizing spectrum. J. Abnorm. Psychol 111, 411–424 (2002). - PubMed
Publication types
MeSH terms
Grants and funding
- UL1 TR000445/TR/NCATS NIH HHS/United States
- P01 HD031921/HD/NICHD NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- U19 HL065962/HL/NHLBI NIH HHS/United States
- R01 HD073342/HD/NICHD NIH HHS/United States
- S10 RR025141/RR/NCRR NIH HHS/United States
- P2C HD042849/HD/NICHD NIH HHS/United States
- RC2 MH089924/MH/NIMH NIH HHS/United States
- R01 HD083613/HD/NICHD NIH HHS/United States
- P50 GM115305/GM/NIGMS NIH HHS/United States
- U01 HG006378/HG/NHGRI NIH HHS/United States
- P50 DA037844/DA/NIDA NIH HHS/United States
- RC2 MH089983/MH/NIMH NIH HHS/United States
- R01 HD092548/HD/NICHD NIH HHS/United States
- R01 DA050721/DA/NIDA NIH HHS/United States
- R01 AA026281/AA/NIAAA NIH HHS/United States
- R01 HD060726/HD/NICHD NIH HHS/United States
- R01 HD074711/HD/NICHD NIH HHS/United States
- P50 AA022537/AA/NIAAA NIH HHS/United States
- K02 AA018755/AA/NIAAA NIH HHS/United States
- RC2 GM092618/GM/NIGMS NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- R01 AA015416/AA/NIAAA NIH HHS/United States
- R01 NS032830/NS/NINDS NIH HHS/United States
- U01 HG004798/HG/NHGRI NIH HHS/United States
- U10 AA008401/AA/NIAAA NIH HHS/United States
- UL1 TR002243/TR/NCATS NIH HHS/United States
- R01 MH120219/MH/NIMH NIH HHS/United States
- R01 DA044283/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

