Label-Free Quantitative Proteomic Analysis of the Global Response to Indole-3-Acetic Acid in Newly Isolated Pseudomonas sp. Strain LY1
- PMID: 34447357
- PMCID: PMC8383072
- DOI: 10.3389/fmicb.2021.694874
Label-Free Quantitative Proteomic Analysis of the Global Response to Indole-3-Acetic Acid in Newly Isolated Pseudomonas sp. Strain LY1
Abstract
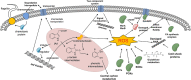
Indole-3-acetic acid (IAA), known as a common plant hormone, is one of the most distributed indole derivatives in the environment, but the degradation mechanism and cellular response network to IAA degradation are still not very clear. The objective of this study was to elucidate the molecular mechanisms of IAA degradation at the protein level by a newly isolated strain Pseudomonas sp. LY1. Label-free quantitative proteomic analysis of strain LY1 cultivated with IAA or citrate/NH4Cl was applied. A total of 2,604 proteins were identified, and 227 proteins have differential abundances in the presence of IAA, including 97 highly abundant proteins and 130 less abundant proteins. Based on the proteomic analysis an IAA degrading (iad) gene cluster in strain LY1 containing IAA transformation genes (organized as iadHABICDEFG), genes of the β-ketoadipate pathway for catechol and protocatechuate degradation (catBCA and pcaABCDEF) were identified. The iadA, iadB, and iadE-disrupted mutants lost the ability to grow on IAA, which confirmed the role of the iad cluster in IAA degradation. Degradation intermediates were analyzed by HPLC, LC-MS, and GC-MS analysis. Proteomic analysis and identified products suggested that multiple degradation pathways existed in strain LY1. IAA was initially transformed to dioxindole-3-acetic acid, which was further transformed to isatin. Isatin was then transformed to isatinic acid or catechol. An in-depth data analysis suggested oxidative stress in strain LY1 during IAA degradation, and the abundance of a series of proteins was upregulated to respond to the stress, including reaction oxygen species (ROS) scavenging, protein repair, fatty acid synthesis, RNA protection, signal transduction, chemotaxis, and several membrane transporters. The findings firstly explained the adaptation mechanism of bacteria to IAA degradation.
Keywords: Pseudomonas sp. LY1; indole-3-acetic acid; microbial degradation; proteomics; stress response.
Copyright © 2021 Zhao, Chen, Sun, Wang, Hu, Guo, Bai and Yu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
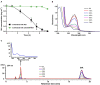
 ) and citrate/NH4Cl (
) and citrate/NH4Cl ( ), respectively. The cell density of resting cells is OD600 = 6.0. The reaction was performed at 30°C 120 rpm. (B) UV scanning of the samples at interval times from IAA transformation reactions. The reactions were performed with resting cells of strain LY1 cultivated with IAA. (C) HPLC analysis of the samples at interval times from IAA transformation reactions. The reactions were performed with resting cells of strain LY1 cultivated with IAA. The signal at 254 nm was recorded. The arrow indicated the UV spectrum of the compound with a retention time of 3.60 min. The mobile phase was 35% (v/v) methanol and 65% (v/v) 0.05% formic acid. The HPLC column was Agilent XBD C18 (4.6 mm × 250 mm, 5 μm). Each value is the mean from three parallel replicates ± SD.
), respectively. The cell density of resting cells is OD600 = 6.0. The reaction was performed at 30°C 120 rpm. (B) UV scanning of the samples at interval times from IAA transformation reactions. The reactions were performed with resting cells of strain LY1 cultivated with IAA. (C) HPLC analysis of the samples at interval times from IAA transformation reactions. The reactions were performed with resting cells of strain LY1 cultivated with IAA. The signal at 254 nm was recorded. The arrow indicated the UV spectrum of the compound with a retention time of 3.60 min. The mobile phase was 35% (v/v) methanol and 65% (v/v) 0.05% formic acid. The HPLC column was Agilent XBD C18 (4.6 mm × 250 mm, 5 μm). Each value is the mean from three parallel replicates ± SD.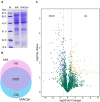




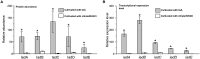

References
-
- Arora P. K., Sharma A., Bae H. (2015). Microbial degradation of indole and its derivatives. J. Chem. 2015:13.
-
- Claus G., Kutzner H. J. (1983). Degradation of indole by Alcaligenes spec. System. Appl. Microbiol. 4 169–180. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

