Transcriptional Profiling and Functional Analysis of N1/N2 Neutrophils Reveal an Immunomodulatory Effect of S100A9-Blockade on the Pro-Inflammatory N1 Subpopulation
- PMID: 34447377
- PMCID: PMC8384118
- DOI: 10.3389/fimmu.2021.708770
Transcriptional Profiling and Functional Analysis of N1/N2 Neutrophils Reveal an Immunomodulatory Effect of S100A9-Blockade on the Pro-Inflammatory N1 Subpopulation
Abstract
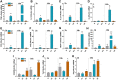
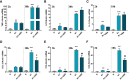
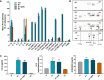
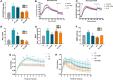
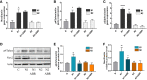
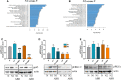
Neutrophils have been classically viewed as a homogenous population. Recently, neutrophils were phenotypically classified into pro-inflammatory N1 and anti-inflammatory N2 sub-populations, but the functional differences between the two subtypes are not completely understood. We aimed to investigate the phenotypic and functional differences between N1 and N2 neutrophils, and to identify the potential contribution of the S100A9 alarmin in neutrophil polarization. We describe distinct transcriptomic profiles and functional differences between N1 and N2 neutrophils. Compared to N2, the N1 neutrophils exhibited: i) higher levels of ROS and oxidative burst, ii) increased activity of MPO and MMP-9, and iii) enhanced chemotactic response. N1 neutrophils were also characterized by elevated expression of NADPH oxidase subunits, as well as activation of the signaling molecules ERK and the p65 subunit of NF-kB. Moreover, we found that the S100A9 alarmin promotes the chemotactic and enzymatic activity of N1 neutrophils. S100A9 inhibition with a specific small-molecule blocker, reduced CCL2, CCL3 and CCL5 chemokine expression and decreased MPO and MMP-9 activity, by interfering with the NF-kB signaling pathway. Together, these findings reveal that N1 neutrophils are pro-inflammatory effectors of the innate immune response. Pharmacological blockade of S100A9 dampens the function of the pro-inflammatory N1 phenotype, promoting the alarmin as a novel target for therapeutic intervention in inflammatory diseases.
Keywords: ABR-238901; N1 neutrophils; N2 neutrophils; RNA-Seq; S100A8/A9; neutrophil chemotaxis; neutrophil polarization.
Copyright © 2021 Mihaila, Ciortan, Macarie, Vadana, Cecoltan, Preda, Hudita, Gan, Jakobsson, Tucureanu, Barbu, Balanescu, Simionescu, Schiopu and Butoi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Denny MF, Yalavarthi S, Zhao W, Thacker SG, Anderson M, Sandy AR, et al. . A Distinct Subset of Proinflammatory Neutrophils Isolated From Patients With Systemic Lupus Erythematosus Induces Vascular Damage and Synthesizes Type I IFNs. J Immunol (2010) 184(6):3284–97. 10.4049/jimmunol.0902199 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

