Multifunctional oncolytic nanoparticles deliver self-replicating IL-12 RNA to eliminate established tumors and prime systemic immunity
- PMID: 34447945
- PMCID: PMC8386348
- DOI: 10.1038/s43018-020-0095-6
Multifunctional oncolytic nanoparticles deliver self-replicating IL-12 RNA to eliminate established tumors and prime systemic immunity
Abstract
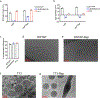
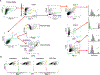
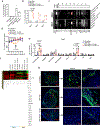
Therapies that synergistically stimulate immunogenic cancer cell death (ICD), inflammation, and immune priming are of great interest for cancer immunotherapy. However, even multi-agent therapies often fail to trigger all of the steps necessary for self-sustaining anti-tumor immunity. Here we describe self-replicating RNAs encapsulated in lipid nanoparticles (LNP-replicons), which combine three key elements: (1) an LNP composition that potently promotes ICD, (2) RNA that stimulates danger sensors in transfected cells, and (3) RNA-encoded IL-12 for modulation of immune cells. Intratumoral administration of LNP-replicons led to high-level expression of IL-12, stimulation of a type I interferon response, and cancer cell ICD, resulting in a highly inflamed tumor microenvironment and priming of systemic anti-tumor immunity. In several mouse models of cancer, a single intratumoral injection of replicon-LNPs eradicated large established tumors, induced protective immune memory, and enabled regression of distal uninjected tumors. LNP-replicons are thus a promising multifunctional single-agent immunotherapeutic.
Figures








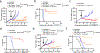


Comment in
-
Pseudovirus for immunotherapy.Nat Cancer. 2020 Sep;1(9):860-861. doi: 10.1038/s43018-020-00107-2. Nat Cancer. 2020. PMID: 35121951 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

