Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations
- PMID: 34450027
- PMCID: PMC8459317
- DOI: 10.1016/j.cell.2021.07.038
Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations
Erratum in
-
Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations.Cell. 2021 Nov 24;184(24):6003-6005. doi: 10.1016/j.cell.2021.11.003. Cell. 2021. PMID: 34822786 Free PMC article. No abstract available.
Abstract
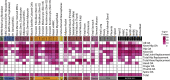
Osteoarthritis affects over 300 million people worldwide. Here, we conduct a genome-wide association study meta-analysis across 826,690 individuals (177,517 with osteoarthritis) and identify 100 independently associated risk variants across 11 osteoarthritis phenotypes, 52 of which have not been associated with the disease before. We report thumb and spine osteoarthritis risk variants and identify differences in genetic effects between weight-bearing and non-weight-bearing joints. We identify sex-specific and early age-at-onset osteoarthritis risk loci. We integrate functional genomics data from primary patient tissues (including articular cartilage, subchondral bone, and osteophytic cartilage) and identify high-confidence effector genes. We provide evidence for genetic correlation with phenotypes related to pain, the main disease symptom, and identify likely causal genes linked to neuronal processes. Our results provide insights into key molecular players in disease processes and highlight attractive drug targets to accelerate translation.
Keywords: drug targets; effector genes; functional genomics; genetic architecture; genome-wide association meta-analysis; osteoarthritis.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.R.G. and J.Z. receive research funding from GlaxoSmithKline. T.R.G. receives research funding from Biogen. U.S., K.S., L. Stefánsdóttir, G.B., S.H.L., U.T., and G. T. are employed by deCODE genetics/Amgen Inc. A.M.V. is a consultant for Zoe Global Ltd. All other authors report no competing interests. All Regeneron Genetics Center banner authors are current employees and/or stockholders of Regeneron Pharmaceuticals.
Figures



Comment in
-
Causality between Ankylosing Spondylitis and osteoarthritis in European ancestry: a bidirectional Mendelian randomization study.Front Immunol. 2024 Feb 6;15:1297454. doi: 10.3389/fimmu.2024.1297454. eCollection 2024. Front Immunol. 2024. PMID: 38380324 Free PMC article.
References
-
- Abdollahi-Roodsaz S., Joosten L.A., Roelofs M.F., Radstake T.R., Matera G., Popa C., van der Meer J.W., Netea M.G., van den Berg W.B. Inhibition of Toll-like receptor 4 breaks the inflammatory loop in autoimmune destructive arthritis. Arthritis Rheum. 2007;56:2957–2967. - PubMed
-
- Aguet F., Barbeira A.N., Bonazzola R., Brown A., Castel S.E., Jo B., Kasela S., Kim-Hellmuth S., Liang Y., Oliva M. The GTEx Consortium atlas of genetic regulatory effects across human tissues. bioRxiv. 2019 doi: 10.1101/787903. - DOI
-
- Alman B.A. The role of hedgehog signalling in skeletal health and disease. Nat. Rev. Rheumatol. 2015;11:552–560. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

