The porphyrin TMPyP4 inhibits elongation during the noncanonical translation of the FTLD/ALS-associated GGGGCC repeat in the C9orf72 gene
- PMID: 34450161
- PMCID: PMC8446798
- DOI: 10.1016/j.jbc.2021.101120
The porphyrin TMPyP4 inhibits elongation during the noncanonical translation of the FTLD/ALS-associated GGGGCC repeat in the C9orf72 gene
Abstract
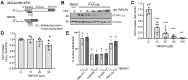
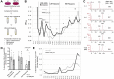
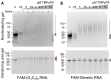
GGGGCC (G4C2) repeat expansion in the C9orf72 gene has been shown to cause frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Dipeptide repeat proteins produced through repeat-associated non-AUG (RAN) translation are recognized as potential drivers for neurodegeneration. Therefore, selective inhibition of RAN translation could be a therapeutic avenue to treat these neurodegenerative diseases. It was previously known that the porphyrin TMPyP4 binds to G4C2 repeat RNA. However, the consequences of this interaction have not been well characterized. Here, we confirmed that TMPyP4 inhibits C9orf72 G4C2 repeat translation in cellular and in in vitro translation systems. An artificial insertion of an AUG codon failed to cancel the translation inhibition, suggesting that TMPyP4 acts downstream of non-AUG translation initiation. Polysome profiling assays also revealed polysome retention on G4C2 repeat RNA, along with inhibition of translation, indicating that elongating ribosomes stall on G4C2 repeat RNA. Urea-resistant interaction between G4C2 repeat RNA and TMPyP4 likely contributes to this ribosome stalling and thus to selective inhibition of RAN translation. Taken together, our data reveal a novel mode of action of TMPyP4 as an inhibitor of G4C2 repeat translation elongation.
Keywords: DPR; G-quadruplex; RAN translation; elongation; frontotemporal dementia; inhibitor; microsatellite; motor neuron disease; repeat expansion; ribosome stalling.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins.J Biol Chem. 2014 Feb 21;289(8):4653-9. doi: 10.1074/jbc.C113.502336. Epub 2013 Dec 26. J Biol Chem. 2014. PMID: 24371143 Free PMC article.
-
eIF5 stimulates the CUG initiation of RAN translation of poly-GA dipeptide repeat protein (DPR) in C9orf72 FTLD/ALS.J Biol Chem. 2024 Mar;300(3):105703. doi: 10.1016/j.jbc.2024.105703. Epub 2024 Jan 30. J Biol Chem. 2024. PMID: 38301895 Free PMC article.
-
Ribosome profiling reveals novel regulation of C9ORF72 GGGGCC repeat-containing RNA translation.RNA. 2022 Feb;28(2):123-138. doi: 10.1261/rna.078963.121. Epub 2021 Nov 30. RNA. 2022. PMID: 34848561 Free PMC article.
-
Unconventional features of C9ORF72 expanded repeat in amyotrophic lateral sclerosis and frontotemporal lobar degeneration.Neurobiol Aging. 2014 Oct;35(10):2421.e1-2421.e12. doi: 10.1016/j.neurobiolaging.2014.04.015. Epub 2014 Apr 19. Neurobiol Aging. 2014. PMID: 24836899 Review.
-
C9orf72 isoforms in Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration.Brain Res. 2016 Sep 15;1647:43-49. doi: 10.1016/j.brainres.2016.04.062. Epub 2016 Apr 29. Brain Res. 2016. PMID: 27134035 Review.
Cited by
-
RNA Dysmetabolism and Repeat-Associated Non-AUG Translation in Frontotemporal Lobar Degeneration/Amyotrophic Lateral Sclerosis due to C9orf72 Hexanucleotide Repeat Expansion.JMA J. 2023 Jan 16;6(1):9-15. doi: 10.31662/jmaj.2022-0160. Epub 2022 Dec 23. JMA J. 2023. PMID: 36793534 Free PMC article. Review.
-
Impaired ribosome-associated quality control of C9orf72 arginine-rich dipeptide-repeat proteins.Brain. 2023 Jul 3;146(7):2897-2912. doi: 10.1093/brain/awac479. Brain. 2023. PMID: 36516294 Free PMC article.
-
G-Quadruplexes Formation by the C9orf72 Nucleotide Repeat Expansion d(GGGGCC)n and Conformation Regulation by Fangchinoline.Molecules. 2023 Jun 9;28(12):4671. doi: 10.3390/molecules28124671. Molecules. 2023. PMID: 37375224 Free PMC article.
-
Proteostasis Deregulation in Neurodegeneration and Its Link with Stress Granules: Focus on the Scaffold and Ribosomal Protein RACK1.Cells. 2022 Aug 19;11(16):2590. doi: 10.3390/cells11162590. Cells. 2022. PMID: 36010666 Free PMC article. Review.
-
FUS regulates RAN translation through modulating the G-quadruplex structure of GGGGCC repeat RNA in C9orf72-linked ALS/FTD.Elife. 2023 Jul 18;12:RP84338. doi: 10.7554/eLife.84338. Elife. 2023. PMID: 37461319 Free PMC article.
References
-
- Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., Kalimo H., Paetau A., Abramzon Y., Remes A.M., Kaganovich A. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. - PMC - PubMed
-
- Gijselinck I., Van Langenhove T., van der Zee J., Sleegers K., Philtjens S., Kleinberger G., Janssens J., Bettens K., Van Cauwenberghe C., Pereson S., Engelborghs S., Sieben A., De Jonghe P., Vandenberghe R., Santens P. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol. 2012;11:54–65. - PubMed
-
- DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., Kouri N., Wojtas A., Sengdy P., Hsiung G.Y., Karydas A. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. - PMC - PubMed
-
- Lee Y.B., Chen H.J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C., Adachi Y., Sardone V., Miller J.W., Smith B.N., Gallo J.M. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. - PMC - PubMed
-
- Mori K., Lammich S., Mackenzie I.R., Forne I., Zilow S., Kretzschmar H., Edbauer D., Janssens J., Kleinberger G., Cruts M., Herms J., Neumann M., Van Broeckhoven C., Arzberger T., Haass C. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125:413–423. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

