Direct Amperometric Sensing of Fish Nodavirus RNA Using Gold Nanoparticle/DNA-Based Bioconjugates
- PMID: 34451396
- PMCID: PMC8398327
- DOI: 10.3390/pathogens10080932
Direct Amperometric Sensing of Fish Nodavirus RNA Using Gold Nanoparticle/DNA-Based Bioconjugates
Abstract
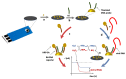
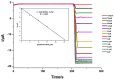
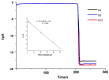
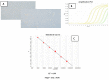
We describe the design of a simple and highly sensitive electrochemical bioanalytical method enabling the direct detection of a conserved RNA region within the capsid protein gene of a fish nodavirus, making use of nanostructured disposable electrodes. To achieve this goal, we select a conserved region within the nodavirus RNA2 segment to design a DNA probe that is tethered to the surface of nanostructured disposable screen-printed electrodes. In a proof-of-principle test, a synthetic RNA sequence is detected based on competitive hybridization between two oligonucleotides (biotinylated reporter DNA and target RNA) complimentary to a thiolated DNA capture probe. The method is further validated using extracted RNA samples obtained from healthy carrier Sparus aurata and clinically infected Dicentrarchus labrax fish specimens. In parallel, the sensitivity of the newly described biosensor is compared with a new real-time RT-PCR protocol. The current differences measured in the negative control and in presence of each concentration of target RNA are used to determine the dynamic range of the assay. We obtain a linear response (R2 = 0.995) over a range of RNA concentrations from 0.1 to 25 pM with a detection limit of 20 fM. The results are in good agreement with the results found by the RT-qPCR. This method provides a promising approach toward a more effective diagnosis and risk assessment of viral diseases in aquaculture.
Keywords: RT-PCR; amperometry; betanodavirus; diagnosis; fish; nanosensors.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Competitive RNA-RNA hybridization-based integrated nanostructured-disposable electrode for highly sensitive determination of miRNAs in cancer cells.Biosens Bioelectron. 2017 May 15;91:40-45. doi: 10.1016/j.bios.2016.12.033. Epub 2016 Dec 13. Biosens Bioelectron. 2017. PMID: 27987409
-
Characterization of the capsid protein gene from a nodavirus strain affecting the Atlantic halibut Hippoglossus hippoglossus and design of an optimal reverse-transcriptase polymerase chain reaction (RT-PCR) detection assay.Dis Aquat Organ. 2000 Jan 14;39(2):79-88. doi: 10.3354/dao039079. Dis Aquat Organ. 2000. PMID: 10715813
-
Real-time RT-PCR detection of betanodavirus in naturally and experimentally infected fish from Spain.J Fish Dis. 2011 Mar;34(3):189-202. doi: 10.1111/j.1365-2761.2010.01227.x. J Fish Dis. 2011. PMID: 21306586
-
Nanoparticle-based lateral flow biosensor for visual detection of fish nervous necrosis virus amplification products.Mol Cell Probes. 2015 Jun;29(3):158-66. doi: 10.1016/j.mcp.2015.03.005. Epub 2015 Mar 20. Mol Cell Probes. 2015. PMID: 25797786
-
A sensitive and label-free impedimetric biosensor based on an adjunct probe.Anal Chim Acta. 2013 May 7;776:11-6. doi: 10.1016/j.aca.2013.03.030. Epub 2013 Mar 21. Anal Chim Acta. 2013. PMID: 23601275 Review.
Cited by
-
A Review of Advanced Impedance Biosensors with Microfluidic Chips for Single-Cell Analysis.Biosensors (Basel). 2021 Oct 22;11(11):412. doi: 10.3390/bios11110412. Biosensors (Basel). 2021. PMID: 34821628 Free PMC article. Review.
-
A Magnetoelectrochemical Bioassay for Highly Sensitive Sensing of Point Mutations in Interleukin-6 Gene Using TMB as a Hybridization Intercalation Indicator.Biosensors (Basel). 2023 Feb 8;13(2):240. doi: 10.3390/bios13020240. Biosensors (Basel). 2023. PMID: 36832006 Free PMC article.
References
-
- Thiery R., Johnson K., Nakai T., Schneemann A., Bonami J., Lightner D. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; Amsterdam, The Netherlands: 2012. Nodaviridae.
LinkOut - more resources
Full Text Sources

