The Structural Biology of Eastern Equine Encephalitis Virus, an Emerging Viral Threat
- PMID: 34451437
- PMCID: PMC8400090
- DOI: 10.3390/pathogens10080973
The Structural Biology of Eastern Equine Encephalitis Virus, an Emerging Viral Threat
Abstract
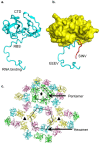
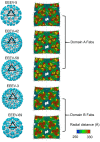
Alphaviruses are arboviruses that cause arthritis and encephalitis in humans. Eastern Equine Encephalitis Virus (EEEV) is a mosquito-transmitted alphavirus that is implicated in severe encephalitis in humans with high mortality. However, limited insights are available into the fundamental biology of EEEV and residue-level details of its interactions with host proteins. In recent years, outbreaks of EEEV have been reported mainly in the United States, raising concerns about public safety. This review article summarizes recent advances in the structural biology of EEEV based mainly on single-particle cryogenic electron microscopy (cryoEM) structures. Together with functional analyses of EEEV and related alphaviruses, these structural investigations provide clues to how EEEV interacts with host proteins, which may open avenues for the development of therapeutics.
Keywords: alphavirus; antibody; assembly; eastern equine encephalitis virus; structure.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources

