Influenza Virus-like Particle (VLP) Vaccines Expressing the SARS-CoV-2 S Glycoprotein, S1, or S2 Domains
- PMID: 34452044
- PMCID: PMC8402567
- DOI: 10.3390/vaccines9080920
Influenza Virus-like Particle (VLP) Vaccines Expressing the SARS-CoV-2 S Glycoprotein, S1, or S2 Domains
Abstract
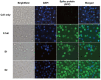
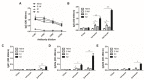
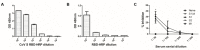
The ongoing severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic had brought disastrous consequences throughout the entire world. While several manufactured vaccines have been approved for emergency use, continuous efforts to generate novel vaccines are needed. In this study, we developed SARS-CoV-2 virus-like particles (VLPs) containing the full length of spike (S) glycoprotein (S full), S1, or S2 together with the influenza matrix protein 1 (M1) as a core protein. Successfully constructed VLPs expressing the S full, S1, and S2 via Sf9 cell transfections were confirmed and characterized by Western blot and transmission electron microscopy (TEM). VLP immunization in mice induced higher levels of spike protein-specific IgG and its subclasses compared to naïve control, with IgG2a being the most predominant subclass. S full and S1 immune sera elicited virus-neutralizing activities, but these were not strong enough to fully inhibit receptor-ligand binding of the SARS-CoV-2. Neutralizing activities were not observed from the S2 VLP immune sera. Overall, our findings revealed that S full or S1 containing VLPs can be developed into effective vaccines.
Keywords: COVID-19; SARS-CoV-2; antibody; neutralization; vaccine; virus-like particle.
Conflict of interest statement
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


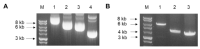


Similar articles
-
Two-Component Nanoparticle Vaccine Displaying Glycosylated Spike S1 Domain Induces Neutralizing Antibody Response against SARS-CoV-2 Variants.mBio. 2021 Oct 26;12(5):e0181321. doi: 10.1128/mBio.01813-21. Epub 2021 Oct 12. mBio. 2021. PMID: 34634927 Free PMC article.
-
Pseudotyping Improves the Yield of Functional SARS-CoV-2 Virus-like Particles (VLPs) as Tools for Vaccine and Therapeutic Development.Int J Mol Sci. 2023 Sep 27;24(19):14622. doi: 10.3390/ijms241914622. Int J Mol Sci. 2023. PMID: 37834067 Free PMC article.
-
A stable platform for the production of virus-like particles pseudotyped with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike protein.Virus Res. 2021 Apr 2;295:198305. doi: 10.1016/j.virusres.2021.198305. Epub 2021 Jan 19. Virus Res. 2021. PMID: 33482242 Free PMC article.
-
Microneedle array delivered recombinant coronavirus vaccines: Immunogenicity and rapid translational development.EBioMedicine. 2020 May;55:102743. doi: 10.1016/j.ebiom.2020.102743. Epub 2020 Apr 2. EBioMedicine. 2020. PMID: 32249203 Free PMC article.
-
An Overview of Recent Developments in the Application of Antigen Displaying Vaccine Platforms: Hints for Future SARS-CoV-2 VLP Vaccines.Vaccines (Basel). 2023 Sep 20;11(9):1506. doi: 10.3390/vaccines11091506. Vaccines (Basel). 2023. PMID: 37766182 Free PMC article. Review.
Cited by
-
Viral vector- and virus-like particle-based vaccines against infectious diseases: A minireview.Heliyon. 2024 Jul 20;10(15):e34927. doi: 10.1016/j.heliyon.2024.e34927. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39144987 Free PMC article. Review.
-
Construction, Characterization, and Application of a Nonpathogenic Virus-like Model for SARS-CoV-2 Nucleocapsid Protein by Phage Display.Toxins (Basel). 2022 Oct 4;14(10):683. doi: 10.3390/toxins14100683. Toxins (Basel). 2022. PMID: 36287952 Free PMC article.
-
Respiratory Viruses and Virus-like Particle Vaccine Development: How Far Have We Advanced?Viruses. 2023 Jan 30;15(2):392. doi: 10.3390/v15020392. Viruses. 2023. PMID: 36851606 Free PMC article. Review.
-
Nanoparticles and Antiviral Vaccines.Vaccines (Basel). 2023 Dec 27;12(1):30. doi: 10.3390/vaccines12010030. Vaccines (Basel). 2023. PMID: 38250843 Free PMC article. Review.
-
Special Issue on 'Coronavirus: Vaccines and Other Therapeutics' (2020-2021).Vaccines (Basel). 2021 Sep 26;9(10):1083. doi: 10.3390/vaccines9101083. Vaccines (Basel). 2021. PMID: 34696191 Free PMC article.
References
-
- van der Hoek L. Human coronaviruses: What do they cause? Antivir. Ther. 2007;12:651–658. - PubMed
-
- Karthik K., Senthilkumar T.M.A., Udhayavel S., Raj G.D. Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19. Hum. Vaccin. Immunother. 2020;16:3055–3060. doi: 10.1080/21645515.2020.1796425. - DOI - PMC - PubMed
-
- WHO WHO Coronavirus (COVID-19) Dashboard. [(accessed on 21 April 2021)]; Available online: https://covid19.who.int/
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

