Detection of Human Papillomavirus Integration in Brain Metastases from Oropharyngeal Tumors by Targeted Sequencing
- PMID: 34452401
- PMCID: PMC8402651
- DOI: 10.3390/v13081536
Detection of Human Papillomavirus Integration in Brain Metastases from Oropharyngeal Tumors by Targeted Sequencing
Abstract
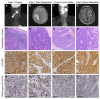
Human papillomavirus (HPV) positive and negative head and neck squamous cell carcinoma (HNSCC) are known to have differential phenotypes, including the incidence and location of metastases. HPV positive (HPV+) HNSCC are more likely to metastasize to distant sites, such as the lung, brain, and skin. Among these locations, metastasis to the brain is a rare event, and little is known about specific risk factors for this phenotype. In this report, we describe two patients who developed brain metastases from HNSCC. Both patient tumors had p16INK4a overexpression, suggesting these tumors were HPV+. This was confirmed after PCR, in situ hybridization, and mass spectrometry detected the presence of HPV type 16 (HPV16) DNA, RNA and protein. To further characterize the presence of HPV16, we used a target enrichment strategy on tumor DNA and RNA to isolate the viral sequences from the brain metastases. Analysis by targeted next generation sequencing revealed that both tumors had the HPV genome integrated into the host genome at known hotspots, 8q24.21 and 14q24.1. Applying a similar target enrichment strategy to a larger cohort of HPV+ HNSCC brain metastases could help to identify biomarkers that can predict metastasis and/or identify novel therapeutic options.
Keywords: DNA target enrichment; HNSCC; HPV; OPSCC; brain metastasis; data-independent acquisition; proteomics; targeted sequencing.
Conflict of interest statement
B.C.S. is a founder and shareholder in Proteome Software, which operates in the field of proteomics. The other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Fakhry C., Westra W.H., Li S., Cmelak A., Ridge J.A., Pinto H., Forastiere A., Gillison M.L. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J. Natl. Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

