Active Components of Commonly Prescribed Medicines Affect Influenza A Virus-Host Cell Interaction: A Pilot Study
- PMID: 34452402
- PMCID: PMC8402715
- DOI: 10.3390/v13081537
Active Components of Commonly Prescribed Medicines Affect Influenza A Virus-Host Cell Interaction: A Pilot Study
Abstract
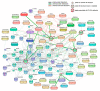
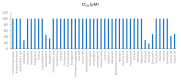
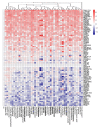
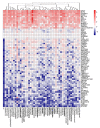
Background: Every year, millions of people are hospitalized and thousands die from influenza A virus (FLUAV) infection. Most cases of hospitalizations and death occur among the elderly. Many of these elderly patients are reliant on medical treatment of underlying chronic diseases, such as arthritis, diabetes, and hypertension. We hypothesized that the commonly prescribed medicines for treatment of underlying chronic diseases can affect host responses to FLUAV infection and thus contribute to the morbidity and mortality associated with influenza. Therefore, the aim of this study was to examine whether commonly prescribed medicines could affect host responses to virus infection in vitro. Methods: We first identified 45 active compounds from a list of commonly prescribed medicines. Then, we constructed a drug-target interaction network and identified the potential implication of these interactions for FLUAV-host cell interplay. Finally, we tested the effect of 45 drugs on the viability, transcription, and metabolism of mock- and FLUAV-infected human retinal pigment epithelial (RPE) cells. Results: In silico drug-target interaction analysis revealed that drugs such as atorvastatin, candesartan, and hydroxocobalamin could target and modulate FLUAV-host cell interaction. In vitro experiments showed that at non-cytotoxic concentrations, these compounds affected the transcription and metabolism of FLUAV- and mock-infected cells. Conclusion: Many commonly prescribed drugs were found to modulate FLUAV-host cell interactions in silico and in vitro and could therefore affect their interplay in vivo, thus contributing to the morbidity and mortality of patients with influenza virus infections.
Keywords: commonly prescribed drugs; drug adverse reaction; influenza virus; virus–host interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

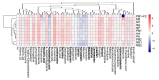
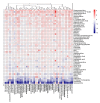
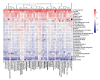
References
-
- Gupta A., Madhavan M.V., Poterucha T.J., DeFilippis E.M., Hennessey J.A., Redfors B., Eckhardt C., Bikdeli B., Platt J., Nalbandian A., et al. Association between antecedent statin use and decreased mortality in hospitalized patients with COVID-19. Nat. Commun. 2021;12:1325. doi: 10.1038/s41467-021-21553-1. - DOI - PMC - PubMed
-
- Anastasina M., Le May N., Bugai A., Fu Y., Soderholm S., Gaelings L., Ohman T., Tynell J., Kyttanen S., Barboric M., et al. Influenza virus NS1 protein binds cellular DNA to block transcription of antiviral genes. Biochim. Biophys. Acta. 2016;1859:1440–1448. doi: 10.1016/j.bbagrm.2016.09.005. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

