Protein Nucleotidylylation in +ssRNA Viruses
- PMID: 34452414
- PMCID: PMC8402628
- DOI: 10.3390/v13081549
Protein Nucleotidylylation in +ssRNA Viruses
Abstract
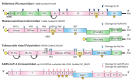
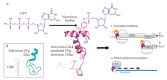
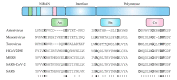
Nucleotidylylation is a post-transcriptional modification important for replication in the picornavirus supergroup of RNA viruses, including members of the Caliciviridae, Coronaviridae, Picornaviridae and Potyviridae virus families. This modification occurs when the RNA-dependent RNA polymerase (RdRp) attaches one or more nucleotides to a target protein through a nucleotidyl-transferase reaction. The most characterized nucleotidylylation target is VPg (viral protein genome-linked), a protein linked to the 5' end of the genome in Caliciviridae, Picornaviridae and Potyviridae. The nucleotidylylation of VPg by RdRp is a critical step for the VPg protein to act as a primer for genome replication and, in Caliciviridae and Potyviridae, for the initiation of translation. In contrast, Coronaviridae do not express a VPg protein, but the nucleotidylylation of proteins involved in replication initiation is critical for genome replication. Furthermore, the RdRp proteins of the viruses that perform nucleotidylylation are themselves nucleotidylylated, and in the case of coronavirus, this has been shown to be essential for viral replication. This review focuses on nucleotidylylation within the picornavirus supergroup of viruses, including the proteins that are modified, what is known about the nucleotidylylation process and the roles that these modifications have in the viral life cycle.
Keywords: RdRp; VPg; calicivirus; coronavirus; nucleotidylylation; picornavirus; potyvirus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
-
- Gorbalenya A.E., Pringle F.M., Zeddam J.L., Luke B.T., Cameron C.E., Kalmakoff J., Hanzlik T.N., Gordon K.H., Ward V.K. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

