Coronavirus Pseudotypes for All Circulating Human Coronaviruses for Quantification of Cross-Neutralizing Antibody Responses
- PMID: 34452443
- PMCID: PMC8402765
- DOI: 10.3390/v13081579
Coronavirus Pseudotypes for All Circulating Human Coronaviruses for Quantification of Cross-Neutralizing Antibody Responses
Abstract
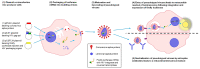
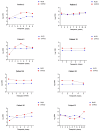
The novel coronavirus SARS-CoV-2 is the seventh identified human coronavirus. Understanding the extent of pre-existing immunity induced by seropositivity to endemic seasonal coronaviruses and the impact of cross-reactivity on COVID-19 disease progression remains a key research question in immunity to SARS-CoV-2 and the immunopathology of COVID-2019 disease. This paper describes a panel of lentiviral pseudotypes bearing the spike (S) proteins for each of the seven human coronaviruses (HCoVs), generated under similar conditions optimized for high titre production allowing a high-throughput investigation of antibody neutralization breadth. Optimal production conditions and most readily available permissive target cell lines were determined for spike-mediated entry by each HCoV pseudotype: SARS-CoV-1, SARS-CoV-2 and HCoV-NL63 best transduced HEK293T/17 cells transfected with ACE2 and TMPRSS2, HCoV-229E and MERS-CoV preferentially entered HUH7 cells, and CHO cells were most permissive for the seasonal betacoronavirus HCoV-HKU1. Entry of ACE2 using pseudotypes was enhanced by ACE2 and TMPRSS2 expression in target cells, whilst TMPRSS2 transfection rendered HEK293T/17 cells permissive for HCoV-HKU1 and HCoV-OC43 entry. Additionally, pseudotype viruses were produced bearing additional coronavirus surface proteins, including the SARS-CoV-2 Envelope (E) and Membrane (M) proteins and HCoV-OC43/HCoV-HKU1 Haemagglutinin-Esterase (HE) proteins. This panel of lentiviral pseudotypes provides a safe, rapidly quantifiable and high-throughput tool for serological comparison of pan-coronavirus neutralizing responses; this can be used to elucidate antibody dynamics against individual coronaviruses and the effects of antibody cross-reactivity on clinical outcome following natural infection or vaccination.
Keywords: COVID-19; SARS-CoV-2; coronavirus; neutralization; pseudotyped virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Characterization of spike S1/S2 processing and entry pathways of lentiviral pseudoviruses bearing seasonal human coronaviruses NL63, 229E, and HKU1 spikes.Microbiol Spectr. 2025 Mar 4;13(3):e0280824. doi: 10.1128/spectrum.02808-24. Epub 2025 Jan 28. Microbiol Spectr. 2025. PMID: 39873512 Free PMC article.
-
Antibody Mediated Immunity to SARS-CoV-2 and Human Coronaviruses: Multiplex Beads Assay and Volumetric Absorptive Microsampling to Generate Immune Repertoire Cartography.Front Immunol. 2021 Jul 27;12:696370. doi: 10.3389/fimmu.2021.696370. eCollection 2021. Front Immunol. 2021. PMID: 34386006 Free PMC article.
-
Mapping SARS-CoV-2 Antibody Epitopes in COVID-19 Patients with a Multi-Coronavirus Protein Microarray.Microbiol Spectr. 2021 Oct 31;9(2):e0141621. doi: 10.1128/Spectrum.01416-21. Epub 2021 Oct 27. Microbiol Spectr. 2021. PMID: 34704808 Free PMC article.
-
An overview on the seven pathogenic human coronaviruses.Rev Med Virol. 2022 Mar;32(2):e2282. doi: 10.1002/rmv.2282. Epub 2021 Aug 2. Rev Med Virol. 2022. PMID: 34339073 Review.
-
Structural proteins of human coronaviruses: what makes them different?Front Cell Infect Microbiol. 2024 Dec 6;14:1458383. doi: 10.3389/fcimb.2024.1458383. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39711780 Free PMC article. Review.
Cited by
-
Characterization of spike S1/S2 processing and entry pathways of lentiviral pseudoviruses bearing seasonal human coronaviruses NL63, 229E, and HKU1 spikes.Microbiol Spectr. 2025 Mar 4;13(3):e0280824. doi: 10.1128/spectrum.02808-24. Epub 2025 Jan 28. Microbiol Spectr. 2025. PMID: 39873512 Free PMC article.
-
Detection of Serum Cross-Reactive Antibodies and Memory Response to SARS-CoV-2 in Prepandemic and Post-COVID-19 Convalescent Samples.J Infect Dis. 2021 Oct 28;224(8):1305-1315. doi: 10.1093/infdis/jiab333. J Infect Dis. 2021. PMID: 34161567 Free PMC article.
-
Glycan masking of a non-neutralising epitope enhances neutralising antibodies targeting the RBD of SARS-CoV-2 and its variants.Front Immunol. 2023 Feb 23;14:1118523. doi: 10.3389/fimmu.2023.1118523. eCollection 2023. Front Immunol. 2023. PMID: 36911730 Free PMC article.
-
Single Ad26.COV2.S booster dose following two doses of BBIBP-CorV vaccine against SARS-CoV-2 infection in adults: Day 28 results of a phase 1/2 open-label trial.Vaccine. 2023 Jul 19;41(32):4648-4657. doi: 10.1016/j.vaccine.2023.06.043. Epub 2023 Jun 15. Vaccine. 2023. PMID: 37344265 Free PMC article. Clinical Trial.
-
Construction of pseudotyped human coronaviruses and detection of pre-existing antibodies in the human population.Biosaf Health. 2024 Sep 3;6(5):279-285. doi: 10.1016/j.bsheal.2024.09.002. eCollection 2024 Oct. Biosaf Health. 2024. PMID: 40078732 Free PMC article.
References
-
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

