Genomic Epidemiology and Evolution of Duck Hepatitis A Virus
- PMID: 34452457
- PMCID: PMC8402860
- DOI: 10.3390/v13081592
Genomic Epidemiology and Evolution of Duck Hepatitis A Virus
Abstract
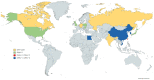
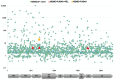
Duck hepatitis A virus (DHAV), an avian picornavirus, causes high-mortality acute disease in ducklings. Among the three serotypes, DHAV-1 is globally distributed, whereas DHAV-2 and DHAV-3 serotypes are chiefly restricted to Southeast Asia. In this study, we analyzed the genomic evolution of DHAV-1 strains using extant GenBank records and genomic sequences of 10 DHAV-1 strains originating from a large disease outbreak in 2004-2005, in Hungary. Recombination analysis revealed intragenotype recombination within DHAV-1 as well as intergenotype recombination events involving DHAV-1 and DHAV-3 strains. The intergenotype recombination occurred in the VP0 region. Diversifying selection seems to act at sites of certain genomic regions. Calculations estimated slightly lower rates of evolution of DHAV-1 (mean rates for individual protein coding regions, 5.6286 × 10-4 to 1.1147 × 10-3 substitutions per site per year) compared to other picornaviruses. The observed evolutionary mechanisms indicate that whole-genome-based analysis of DHAV strains is needed to better understand the emergence of novel strains and their geographical dispersal.
Keywords: Hungary; duck hepatitis A virus; recombination.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Hisham I., Ellakany H.F., Selim A.A., Abdalla M.A.M., El-Abideen M.A.Z., Kilany W.H., Ali A., Elbestawy A.R. Comparative pathogenicity of duck hepatitis A virus-1 isolates in experimentally infected Pekin and Muscovy ducklings. Front. Vet. Sci. 2020;7:234. doi: 10.3389/fvets.2020.00234. - DOI - PMC - PubMed
-
- Levine P.P., Fabricant J. A hitherto-undescribed virus disease of ducks in North America. Cornell Vet. 1950;40:71–86.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

