Adeno-Associated Vector-Delivered CRISPR/ Sa Cas9 System Reduces Feline Leukemia Virus Production In Vitro
- PMID: 34452500
- PMCID: PMC8402633
- DOI: 10.3390/v13081636
Adeno-Associated Vector-Delivered CRISPR/ Sa Cas9 System Reduces Feline Leukemia Virus Production In Vitro
Abstract
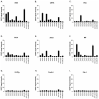
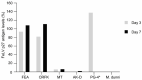
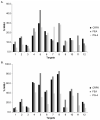
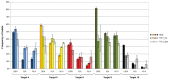
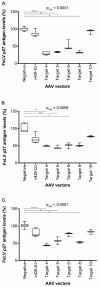
Feline leukemia virus (FeLV) is a retrovirus of cats worldwide. High viral loads are associated with progressive infection and the death of the host, due to FeLV-associated disease. In contrast, low viral loads, an effective immune response, and a better clinical outcome can be observed in cats with regressive infection. We hypothesize that by lowering viral loads in progressively infected cats, using CRISPR/SaCas9-assisted gene therapy, the cat's immune system may be permitted to direct the infection towards a regressive outcome. In a step towards this goal, the present study evaluates different adeno-associated vectors (AAVs) for their competence in delivering a gene editing system into feline cells, followed by investigations of the CRISPR/SaCas9 targeting efficiency for different sites within the FeLV provirus. Nine natural AAV serotypes, two AAV hybrid strains, and Anc80L65, an in silico predicted AAV ancestor, were tested for their potential to infect different feline cell lines and feline primary cells. AAV-DJ revealed superior infection efficiency and was thus employed in subsequent transduction experiments. The introduction of double-strand breaks, using the CRISPR/SaCas9 system targeting 12 selected FeLV provirus sites, was confirmed by T7 endonuclease 1 (T7E1), as well as Tracking of Indels by Decomposition (TIDE) analysis. The highest percentage (up to 80%) of nonhomologous end-joining (NHEJ) was found in the highly conserved gag and pol regions. Subsequent transduction experiments, using AAV-DJ, confirmed indel formation and showed a significant reduction in FeLV p27 antigen for some targets. The targeting of the FeLV provirus was efficient when using the CRISPR/SaCas9 approach in vitro. Whether the observed extent of provirus targeting will be sufficient to provide progressively FeLV-infected cats with the means to overcome the infection needs to be further investigated in vivo.
Keywords: AAV; CRISPR; FeLV; SaCas9; cat; feline leukemia virus; gene editing; provirus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






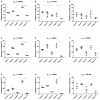
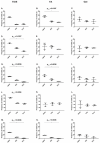

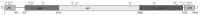
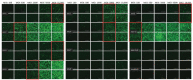
References
-
- UNAIDS Global HIV & AIDS Statistics–2020 Fact Sheet. [(accessed on 2 June 2021)];2021 Available online: https://www.unaids.org/en/resources/fact-sheet.
-
- Studer N., Lutz H., Saegerman C., Gönczi E., Meli M.L., Boo G., Hartmann K., Hosie M.J., Moestl K., Tasker S., et al. Pan-European Study on the Prevalence of the Feline Leukaemia Virus Infection—Reported by the European Advisory Board on Cat Diseases (ABCD Europe) Viruses. 2019;11:993. doi: 10.3390/v11110993. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

