Prevalence of Hantaviruses Harbored by Murid Rodents in Northwestern Ukraine and Discovery of a Novel Puumala Virus Strain
- PMID: 34452504
- PMCID: PMC8402871
- DOI: 10.3390/v13081640
Prevalence of Hantaviruses Harbored by Murid Rodents in Northwestern Ukraine and Discovery of a Novel Puumala Virus Strain
Abstract
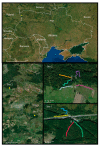
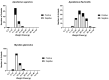
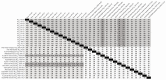
In Europe, two species of hantaviruses, Puumala orthohantavirus (PUUV) and Dobrava orthohantavirus (DOBV), cause hemorrhagic fever with renal syndrome in humans. The rodent reservoirs for these viruses are common throughout Ukraine, and hence, the goal of this study was to identify the species and strains of hantaviruses circulating in this region. We conducted surveillance of small rodent populations in a rural region in northwestern Ukraine approximately 30 km from Poland. From the 424 small mammals captured, we identified nine species, of which the most abundant were Myodes glareolus, the bank vole (45%); Apodemus flavicollis, the yellow-necked mouse (29%); and Apodemus agrarius, the striped field mouse (14.6%) Using an indirect immunofluorescence assay, 15.7%, 20.5%, and 33.9% of the sera from M. glareolus, A. glareolus, and A. flavicollis were positive for hantaviral antibodies, respectively. Additionally, we detected antibodies to the hantaviral antigen in one Microtus arvalis, one Mus musculus, and one Sorex minutus. We screened the lung tissue for hantaviral RNA using next-generation sequencing and identified PUUV sequences in 25 small mammals, including 23 M. glareolus, 1 M. musculus, and 1 A. flavicollis, but we were unable to detect DOBV sequences in any of our A. agrarius specimens. The percent identity matrix and Bayesian phylogenetic analyses of the S-segment of PUUV from 14 M. glareolus lungs suggest the highest similarity (92-95% nucleotide or 99-100% amino acid) with the Latvian lineage. This new genetic information will contribute to future molecular surveillance of human cases in Ukraine.
Keywords: Puumala orthohantavirus; Ukraine; field survey; next generation sequencing.
Conflict of interest statement
I.K. and O.N. were employed by Black and Veatch.
Figures



References
-
- Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Adriaenssens E.M., Dempsey D.M., Dutilh B.E., Harrach B., Harrison R.L., Hendrickson R.C., et al. Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses (2020) Arch. Virol. 2020;165:2737–2748. doi: 10.1007/s00705-020-04752-x. - DOI - PubMed
-
- Avsic-Zupanc T., Nemirov K., Petrovec M., Trilar T., Poljak M., Vaheri A., Plyusnin A. Genetic analysis of wild-type Dobrava hantavirus in Slovenia: Co-existence of two distinct genetic lineages within the same natural focus. J. Gen. Virol. 2000;81:1747–1755. doi: 10.1099/0022-1317-81-7-1747. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

