Serological Detection of SARS-CoV-2 Antibodies in Naturally-Infected Mink and Other Experimentally-Infected Animals
- PMID: 34452513
- PMCID: PMC8402807
- DOI: 10.3390/v13081649
Serological Detection of SARS-CoV-2 Antibodies in Naturally-Infected Mink and Other Experimentally-Infected Animals
Abstract
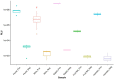
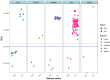
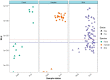
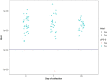
The recent emergence of SARS-CoV-2 in humans from a yet unidentified animal reservoir and the capacity of the virus to naturally infect pets, farmed animals and potentially wild animals has highlighted the need for serological surveillance tools. In this study, the luciferase immunoprecipitation systems (LIPS), employing the spike (S) and nucleocapsid proteins (N) of SARS-CoV-2, was used to examine the suitability of the assay for antibody detection in different animal species. Sera from SARS-CoV-2 naturally-infected mink (n = 77), SARS-CoV-2 experimentally-infected ferrets, fruit bats and hamsters and a rabbit vaccinated with a purified spike protein were examined for antibodies using the SARS-CoV-2 N and/or S proteins. From comparison with the known neutralization status of the serum samples, statistical analyses including calculation of the Spearman rank-order-correlation coefficient and Cohen's kappa agreement were used to interpret the antibody results and diagnostic performance. The LIPS immunoassay robustly detected the presence of viral antibodies in naturally infected SARS-CoV-2 mink, experimentally infected ferrets, fruit bats and hamsters as well as in an immunized rabbit. For the SARS-CoV-2-LIPS-S assay, there was a good level of discrimination between the positive and negative samples for each of the five species tested with 100% agreement with the virus neutralization results. In contrast, the SARS-CoV-2-LIPS-N assay did not consistently differentiate between SARS-CoV-2 positive and negative sera. This study demonstrates the suitability of the SARS-CoV-2-LIPS-S assay for the sero-surveillance of SARS-CoV-2 infection in a range of animal species.
Keywords: SARS-CoV-2; animal; luciferase immunoprecipitation systems; mink; nucleocapsid; sera; spike.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The Luciferase Immunoprecipitation System (LIPS) Targeting the Spike Protein of SARS-CoV-2 Is More Accurate than Nucleoprotein-Based LIPS and ELISAs for Mink Serology.Transbound Emerg Dis. 2023 Feb 22;2023:1318901. doi: 10.1155/2023/1318901. eCollection 2023. Transbound Emerg Dis. 2023. PMID: 40303713 Free PMC article.
-
SARS-CoV-2 Antibody Testing in Health Care Workers: A Comparison of the Clinical Performance of Three Commercially Available Antibody Assays.Microbiol Spectr. 2021 Oct 31;9(2):e0039121. doi: 10.1128/Spectrum.00391-21. Epub 2021 Sep 29. Microbiol Spectr. 2021. PMID: 34585976 Free PMC article.
-
Seroprevalence of SARS-CoV-2 (COVID-19) exposure in pet cats and dogs in Minnesota, USA.Virulence. 2021 Dec;12(1):1597-1609. doi: 10.1080/21505594.2021.1936433. Virulence. 2021. PMID: 34125647 Free PMC article.
-
Transmission dynamics and susceptibility patterns of SARS-CoV-2 in domestic, farmed and wild animals: Sustainable One Health surveillance for conservation and public health to prevent future epidemics and pandemics.Transbound Emerg Dis. 2022 Sep;69(5):2523-2543. doi: 10.1111/tbed.14356. Epub 2021 Nov 9. Transbound Emerg Dis. 2022. PMID: 34694705 Free PMC article. Review.
-
Protein-based lateral flow assays for COVID-19 detection.Protein Eng Des Sel. 2021 Feb 15;34:gzab010. doi: 10.1093/protein/gzab010. Protein Eng Des Sel. 2021. PMID: 33991088 Free PMC article. Review.
Cited by
-
Development of a highly sensitive Gaussia luciferase immunoprecipitation assay for the detection of antibodies against African swine fever virus.Front Cell Infect Microbiol. 2022 Sep 14;12:988355. doi: 10.3389/fcimb.2022.988355. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36189357 Free PMC article.
-
The Luciferase Immunoprecipitation System (LIPS) Targeting the Spike Protein of SARS-CoV-2 Is More Accurate than Nucleoprotein-Based LIPS and ELISAs for Mink Serology.Transbound Emerg Dis. 2023 Feb 22;2023:1318901. doi: 10.1155/2023/1318901. eCollection 2023. Transbound Emerg Dis. 2023. PMID: 40303713 Free PMC article.
-
Dynamics of influenza transmission in vampire bats revealed by longitudinal monitoring and a large-scale anthropogenic perturbation.Sci Adv. 2025 Feb 7;11(6):eads1267. doi: 10.1126/sciadv.ads1267. Epub 2025 Feb 5. Sci Adv. 2025. PMID: 39908385 Free PMC article.
-
SARS-CoV-2 in a Mink Farm in Italy: Case Description, Molecular and Serological Diagnosis by Comparing Different Tests.Viruses. 2022 Aug 8;14(8):1738. doi: 10.3390/v14081738. Viruses. 2022. PMID: 36016360 Free PMC article.
-
Variations in Cell Surface ACE2 Levels Alter Direct Binding of SARS-CoV-2 Spike Protein and Viral Infectivity: Implications for Measuring Spike Protein Interactions with Animal ACE2 Orthologs.J Virol. 2022 Sep 14;96(17):e0025622. doi: 10.1128/jvi.00256-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000847 Free PMC article.
References
-
- Aguiló-Gisbert J., Padilla-Blanco M., Lizana V., Maiques E., Muñoz-Baquero M., Chillida-Martínez E., Cardells J., Rubio-Guerri C. First description of SARS-CoV-2 infection in two feral American mink (Neovison vison) caught in the wild. Animals. 2021;11:1422. doi: 10.3390/ani11051422. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

