RNA and Sugars, Unique Properties of Bacteriophages Infecting Multidrug Resistant Acinetobacter radioresistens Strain LH6
- PMID: 34452516
- PMCID: PMC8402811
- DOI: 10.3390/v13081652
RNA and Sugars, Unique Properties of Bacteriophages Infecting Multidrug Resistant Acinetobacter radioresistens Strain LH6
Abstract
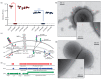
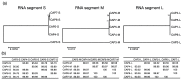
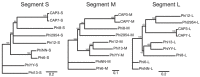
Bacteriophages (phages) are predicted to be the most ubiquitous biological entity on earth, and yet, there are still vast knowledge gaps in our understanding of phage diversity and phage-host interactions. Approximately one hundred Acinetobacter-infecting DNA viruses have been identified, and in this report, we describe eight more. We isolated two typical dsDNA lytic podoviruses (CAP1-2), five unique dsRNA lytic cystoviruses (CAP3-7), and one dsDNA lysogenic siphovirus (SLAP1), all capable of infecting the multidrug resistant isolate Acinetobacter radioresistens LH6. Using transmission electron microscopy, bacterial mutagenesis, phage infectivity assays, carbohydrate staining, mass-spectrometry, genomic sequencing, and comparative studies, we further characterized these phages. Mutation of the LH6 initiating glycosyltransferase homolog, PglC, necessary for both O-linked glycoprotein and capsular polysaccharide (CPS) biosynthesis, prevented infection by the lytic podovirus CAP1, while mutation of the pilin protein, PilA, prevented infection by CAP3, representing the lytic cystoviruses. Genome sequencing of the three dsRNA segments of the isolated cystoviruses revealed low levels of homology, but conserved synteny with the only other reported cystoviruses that infect Pseudomonas species. In Pseudomonas, the cystoviruses are known to be enveloped phages surrounding their capsids with the inner membrane from the infected host. To characterize any membrane-associated glycoconjugates in the CAP3 cystovirus, carbohydrate staining was used to identify a low molecular weight lipid-linked glycoconjugate subsequently identified by mutagenesis and mass-spectrometry as bacterial lipooligosaccharide. Together, this study demonstrates the isolation of new Acinetobacter-infecting phages and the determination of their cell receptors. Further, we describe the genomes of a new genus of Cystoviruses and perform an initial characterization of membrane-associated glycoconjugates.
Keywords: Acinetobacter; bacteriophages; capsular polysaccharides; lipooligosaccharides; pilin; segmented RNA viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Diversity and Current Classification of dsRNA Bacteriophages.Viruses. 2023 Oct 25;15(11):2154. doi: 10.3390/v15112154. Viruses. 2023. PMID: 38005832 Free PMC article. Review.
-
Recognition of six additional cystoviruses: Pseudomonas virus phi6 is no longer the sole species of the family Cystoviridae.Arch Virol. 2018 Apr;163(4):1117-1124. doi: 10.1007/s00705-017-3679-4. Epub 2017 Dec 19. Arch Virol. 2018. PMID: 29260329
-
New enveloped dsRNA phage from freshwater habitat.J Gen Virol. 2015 May;96(Pt 5):1180-1189. doi: 10.1099/vir.0.000063. Epub 2015 Jan 22. J Gen Virol. 2015. PMID: 25614591
-
The characteristics and genome analysis of vB_ApiP_XC38, a novel phage infecting Acinetobacter pittii.Virus Genes. 2020 Aug;56(4):498-507. doi: 10.1007/s11262-020-01766-0. Epub 2020 May 25. Virus Genes. 2020. PMID: 32449140
-
RNA Packaging in the Cystovirus Bacteriophages: Dynamic Interactions during Capsid Maturation.Int J Mol Sci. 2022 Feb 28;23(5):2677. doi: 10.3390/ijms23052677. Int J Mol Sci. 2022. PMID: 35269819 Free PMC article. Review.
Cited by
-
Diversity and Current Classification of dsRNA Bacteriophages.Viruses. 2023 Oct 25;15(11):2154. doi: 10.3390/v15112154. Viruses. 2023. PMID: 38005832 Free PMC article. Review.
-
Bacteriophages and their unique components provide limitless resources for exploitation.Front Microbiol. 2024 Feb 6;15:1342544. doi: 10.3389/fmicb.2024.1342544. eCollection 2024. Front Microbiol. 2024. PMID: 38380101 Free PMC article. No abstract available.
-
Identification of receptor-binding protein and host receptor of non-lytic dsRNA phage phiNY.Microbiol Spectr. 2024 Oct 22;12(12):e0146724. doi: 10.1128/spectrum.01467-24. Online ahead of print. Microbiol Spectr. 2024. PMID: 39436121 Free PMC article.
References
-
- WHO . WHO Model List of Essential Medicines. Volume 20 WHO; Geneva, Switzerland: 2017.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

