Persistence of Multiple Paramyxoviruses in a Closed Captive Colony of Fruit Bats (Eidolon helvum)
- PMID: 34452523
- PMCID: PMC8402880
- DOI: 10.3390/v13081659
Persistence of Multiple Paramyxoviruses in a Closed Captive Colony of Fruit Bats (Eidolon helvum)
Abstract
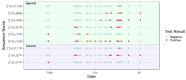
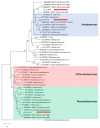
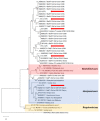
Bats have been identified as the natural hosts of several emerging zoonotic viruses, including paramyxoviruses, such as Hendra and Nipah viruses, that can cause fatal disease in humans. Recently, African fruit bats with populations that roost in or near urban areas have been shown to harbour a great diversity of paramyxoviruses, posing potential spillover risks to public health. Understanding the circulation of these viruses in their reservoir populations is essential to predict and prevent future emerging diseases. Here, we identify a high incidence of multiple paramyxoviruses in urine samples collected from a closed captive colony of circa 115 straw-coloured fruit bats (Eidolon helvum). The sequences detected have high nucleotide identities with those derived from free ranging African fruit bats and form phylogenetic clusters with the Henipavirus genus, Pararubulavirus genus and other unclassified paramyxoviruses. As this colony had been closed for 5 years prior to this study, these results indicate that within-host paramyxoviral persistence underlies the role of bats as reservoirs of these viruses.
Keywords: Henipavirus; Pararubulavirus; Pteropodidae; chiroptera; longitudinal study.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Suu-Ire R., Begeman L., Banyard A.C., Breed A.C., Drosten C., Eggerbauer E., Freuling C.M., Gibson L., Goharriz H., Horton D.L., et al. Pathogenesis of bat rabies in a natural reservoir: Comparative susceptibility of the straw-colored fruit bat (Eidolon helvum) to three strains of Lagos bat virus. PLoS Negl. Trop. Dis. 2018;12:e0006311. doi: 10.1371/journal.pntd.0006311. - DOI - PMC - PubMed
-
- Luis A.D., Hayman D.T.S., O’shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R.C., Mills J.N., Willis C.K.R., Timonin M.E., Cunningham A.A., et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc. Biol. Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

