In vitro data suggest that Indian delta variant B.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion
- PMID: 34453338
- PMCID: PMC8652796
- DOI: 10.1111/all.15065
In vitro data suggest that Indian delta variant B.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion
Abstract
Background: Emerged mutations can be attributed to increased transmissibility of the B.1.617 and B.1.36 Indian delta variants of SARS-CoV-2, most notably substitutions L452R/E484Q and N440K, respectively, which occur in the receptor-binding domain (RBD) of the Spike (S) fusion glycoprotein.
Objective: We aimed to assess the effects of mutations L452R/E484Q and N440K (as well as the previously studied mutation E484K present in variants B.1.351 and P.1) on the affinity of RBD for ACE2, SARS-CoV-2 main receptor. We also aimed to assess the ability of antibodies induced by natural infection or by immunization with BNT162b2 mRNA vaccine to recognize the mutated versions of the RBD, as well as blocking the interaction RBD-ACE2, an important surrogate readout for virus neutralization.
Methods: To this end, we produced recombinant wild-type RBD, as well as RBD containing each of the mutations L452R/E484Q, N440K, or E484K (the latest present in variants of concern B.1.351 and P.1), as well as the ectodomain of ACE2. Using Biolayer Interferometry (BLI), we measured the binding affinity of RBD for ACE2 and the ability of sera from COVID-19 convalescent donors or subjects immunized with BNT162b2 mRNA vaccine to block this interaction. Finally, we correlated these results with total anti-RBD IgG titers measured from the same sera by direct ELISA.
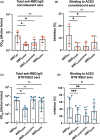
Results: The binding assays showed L452R/E484Q double-mutant RBD to interact with ACE2 with higher affinity (KD = 4.6 nM) than wild-type (KD = 21.3 nM) or single mutants N440K (KD = 9.9 nM) and E484K (KD = 19.7 nM) RBDs. Meanwhile, the anti-RBD IgG titration resulted in lower recognition of mutants E484K and L452R/E484Q by infection-induced antibodies, whereas only mutant E484K was recognized less by antibodies induced by vaccination. More interestingly, sera from convalescent as well as immunized subjects showed reduced ability to block the interaction between ACE2 and RBD mutants E484K and L452R/E484Q, as shown by the inhibition assays.
Conclusion: Our data suggest that the newly emerged SARS-CoV-2 variant B.1.617, as well as the better-studied variants B.1.351 and P.1 (all containing a mutation at position E484) display increased transmissibility both due to their higher affinity for the cell receptor ACE2 and their ability to partially bypass immunity generated against the wild-type virus. For variant B.1.36 (with a point mutation at position N440), only increased affinity seems to play a role.
Keywords: RBD; SARS-CoV-2; affinity; neutralization; vaccine.
© 2021 The Authors. Allergy published by European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
M. F. Bachmann is a board member of Saiba AG, involved in the development of RBD‐CuMV, a vaccine against COVID‐19. All other authors declare no conflict of interest.
Figures


References
-
- Jangra S, Ye C, Rathnasinghe R, et al. The E484K mutation in the SARS‐CoV‐2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post‐vaccination sera. medRxiv 2021.
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

