Gene signatures predict biochemical recurrence-free survival in primary prostate cancer patients after radical therapy
- PMID: 34453418
- PMCID: PMC8446568
- DOI: 10.1002/cam4.4092
Gene signatures predict biochemical recurrence-free survival in primary prostate cancer patients after radical therapy
Abstract
Background: This study evaluated the predictive value of gene signatures for biochemical recurrence (BCR) in primary prostate cancer (PCa) patients.
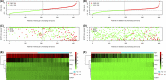
Methods: Clinical features and gene expression profiles of PCa patients were attained from Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) datasets, which were further classified into a training set (n = 419), a validation set (n = 403). The least absolute shrinkage and selection operator Cox (LASSO-Cox) method was used to select discriminative gene signatures in training set for biochemical recurrence-free survival (BCRFS). Selected gene signatures established a risk score system. Univariate and multivariate analyses of prognostic factors about BCRFS were performed using the Cox proportional hazards regression models. A nomogram based on multivariate analysis was plotted to facilitate clinical application. Kyoto Encyclopedia of Gene and Genomes (KEGG) and Gene Ontology (GO) analyses were then executed for differentially expressed genes (DEGs).
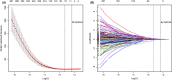
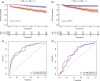
Results: Notably, the risk score could significantly identify BCRFS by time-dependent receiver operating characteristic (t-ROC) curves in the training set (3-year area under the curve (AUC) = 0.820, 5-year AUC = 0.809) and the validation set (3-year AUC = 0.723, 5-year AUC = 0.733).
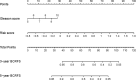
Conclusions: Clinically, the nomogram model, which incorporates Gleason score and the risk score, could effectively predict BCRFS and potentially be utilized as a useful tool for the screening of BCRFS in PCa.
Keywords: LASSO-Cox regression; biochemical recurrence-free survival; gene signature; primary prostate cancer; radical therapy.
© 2021 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors confirm that there are no conflicts of interest.
Figures


References
-
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: Cancer J Clin. 2020;70(1):7‐30. - PubMed
-
- Mottet N, Bellmunt J, Bolla M, et al. EAU‐ESTRO‐SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017;71(4):618‐629. - PubMed
-
- Kupelian PA, Mahadevan A, Reddy CA, et al. Use of different definitions of biochemical failure after external beam radiotherapy changes conclusions about relative treatment efficacy for localized prostate cancer. Urology. 2006;68(3):593‐598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- XDB01030200/Strategic Priority Research Program of Chinese Academy of Sciences
- XDB32030200/Strategic Priority Research Program of Chinese Academy of Sciences
- 2017YFA0205200/National Key R&D Program of China
- 7182109/Natural Science Foundation of Beijing Municipality
- Z200027/Natural Science Foundation of Beijing Municipality
- 201902075003/Ministry of Education of the People's Republic of China
- 81922040/National Natural Science Foundation of China
- 81930053/National Natural Science Foundation of China
- 2017YFA0205200/Ministry of Science and Technology of the People's Republic of China
- 2019136/The Youth Innovation Promotion Association CAS
- QYZDJ-SSW-JSC005/Chinese Academy of Sciences
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

