Prediction of protein assemblies, the next frontier: The CASP14-CAPRI experiment
- PMID: 34453465
- PMCID: PMC8616814
- DOI: 10.1002/prot.26222
Prediction of protein assemblies, the next frontier: The CASP14-CAPRI experiment
Abstract
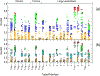
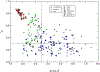
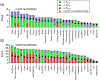
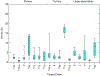
We present the results for CAPRI Round 50, the fourth joint CASP-CAPRI protein assembly prediction challenge. The Round comprised a total of twelve targets, including six dimers, three trimers, and three higher-order oligomers. Four of these were easy targets, for which good structural templates were available either for the full assembly, or for the main interfaces (of the higher-order oligomers). Eight were difficult targets for which only distantly related templates were found for the individual subunits. Twenty-five CAPRI groups including eight automatic servers submitted ~1250 models per target. Twenty groups including six servers participated in the CAPRI scoring challenge submitted ~190 models per target. The accuracy of the predicted models was evaluated using the classical CAPRI criteria. The prediction performance was measured by a weighted scoring scheme that takes into account the number of models of acceptable quality or higher submitted by each group as part of their five top-ranking models. Compared to the previous CASP-CAPRI challenge, top performing groups submitted such models for a larger fraction (70-75%) of the targets in this Round, but fewer of these models were of high accuracy. Scorer groups achieved stronger performance with more groups submitting correct models for 70-80% of the targets or achieving high accuracy predictions. Servers performed less well in general, except for the MDOCKPP and LZERD servers, who performed on par with human groups. In addition to these results, major advances in methodology are discussed, providing an informative overview of where the prediction of protein assemblies currently stands.
Keywords: CAPRI; CASP; blind prediction; docking; oligomeric state; protein assemblies; protein complexes; protein docking; protein-protein interaction; template-based modeling.
© 2021 Wiley Periodicals LLC.
Figures


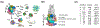
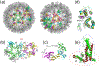

References
-
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. Journal of molecular biology 1999;285(5):2177–2198. - PubMed
-
- Dey S, Pal A, Chakrabarti P, Janin J. The subunit interfaces of weakly associated homodimeric proteins. Journal of molecular biology 2010;398(1):146–160. - PubMed
-
- Ponstingl H, Kabir T, Gorse D, Thornton JM. Morphological aspects of oligomeric protein structures. Progress in biophysics and molecular biology 2005;89(1):9–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R35 GM136409/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 GM074255/GM/NIGMS NIH HHS/United States
- R01 GM109980/GM/NIGMS NIH HHS/United States
- 10748/CRUK_/Cancer Research UK/United Kingdom
- R21 GM127952/GM/NIGMS NIH HHS/United States
- R35 GM118078/GM/NIGMS NIH HHS/United States
- R35 GM124952/GM/NIGMS NIH HHS/United States
- MRC_/Medical Research Council/United Kingdom
- R01 HL142301/HL/NHLBI NIH HHS/United States
- FC001003/CRUK_/Cancer Research UK/United Kingdom
- T32 GM132024/GM/NIGMS NIH HHS/United States
- RM1 GM135136/GM/NIGMS NIH HHS/United States
- R01 GM078221/GM/NIGMS NIH HHS/United States
- R01 GM123055/GM/NIGMS NIH HHS/United States
- R01 GM133840/GM/NIGMS NIH HHS/United States
- R01 GM093123/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

