1H, 13C and 15N assignment of stem-loop SL1 from the 5'-UTR of SARS-CoV-2
- PMID: 34453696
- PMCID: PMC8401371
- DOI: 10.1007/s12104-021-10047-2
1H, 13C and 15N assignment of stem-loop SL1 from the 5'-UTR of SARS-CoV-2
Abstract
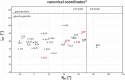
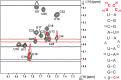
The stem-loop (SL1) is the 5'-terminal structural element within the single-stranded SARS-CoV-2 RNA genome. It is formed by nucleotides 7-33 and consists of two short helical segments interrupted by an asymmetric internal loop. This architecture is conserved among Betacoronaviruses. SL1 is present in genomic SARS-CoV-2 RNA as well as in all subgenomic mRNA species produced by the virus during replication, thus representing a ubiquitous cis-regulatory RNA with potential functions at all stages of the viral life cycle. We present here the 1H, 13C and 15N chemical shift assignment of the 29 nucleotides-RNA construct 5_SL1, which denotes the native 27mer SL1 stabilized by an additional terminal G-C base-pair.
Keywords: 5'-UTR; COVID19-NMR; SARS-CoV-2; SL1; Solution NMR spectroscopy.
© 2021. The Author(s).
Conflict of interest statement
The authors declare the following competing financial interest(s): Daniel Mathieu is an employee of Bruker BioSpin.
Figures

References
-
- Bax A, Vuister GW, Grzesiek S, et al. Nuclear magnetic resonance, Part C. Cambridge: Academic Press; 1994. Measurement of homo- and heteronuclear J couplings from quantitative J correlation; pp. 79–105. - PubMed
-
- Bermel W, Bertini I, Felli IC, et al. 13 C-detected protonless NMR spectroscopy of proteins in solution. Prog Nucl Magn Reson Spectrosc. 2006;48:25–45. doi: 10.1016/j.pnmrs.2005.09.002. - DOI
-
- Bodenhausen G, Ruben DJ. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett. 1980;69:185–189. doi: 10.1016/0009-2614(80)80041-8. - DOI
-
- Breeze AL. Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog Nucl Magn Reson Spectrosc. 2000;4:323–372. doi: 10.1016/S0079-6565(00)00020-0. - DOI
MeSH terms
Substances
Grants and funding
- BMRZ/hessisches ministerium für wissenschaft und kunst
- IWB-EFRE-programme 20007375/hessisches ministerium für wissenschaft und kunst
- CRC 902/deutsche forschungsgemeinschaft
- CLIC/deutsche forschungsgemeinschaft
- SPP2002/deutsche forschungsgemeinschaft
- SPP1879/deutsche forschungsgemeinschaft
- TRR 267/deutsche forschungsgemeinschaft
- FOR2509/deutsche forschungsgemeinschaft
- 277478796/deutsche forschungsgemeinschaft
- 277479031/deutsche forschungsgemeinschaft
- 392682309/deutsche forschungsgemeinschaft
- 452632086/deutsche forschungsgemeinschaft
- 70653611/deutsche forschungsgemeinschaft
- Goethe Corona Fonds/goethe-universität frankfurt am main
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

