Establishment of human induced trophoblast stem-like cells from term villous cytotrophoblasts
- PMID: 34454392
- PMCID: PMC8551050
- DOI: 10.1016/j.scr.2021.102507
Establishment of human induced trophoblast stem-like cells from term villous cytotrophoblasts
Abstract
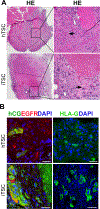
Human trophoblast stem cells (hTSC) can be isolated from first trimester placenta but not from term placenta. Here we demonstrate that villous cytotrophoblasts (vCTB) from term placenta can be reprogrammed into induced trophoblastic stem-like cells (iTSC) by introducing sets of transcription factors. The iTSCs express TSC markers such as GATA3, TEAD4 and ELF5, and are multipotent, validated by their differentiation into both extravillous trophoblasts (EVT) and syncytiotrophoblasts (STB) in vitro and in vivo. The iTSC can be passaged indefinitely in vitro without slowing of growth. The transcriptome profile of these cells closely resembles the profile of hTSC isolated from first trimester placentae but different from the term placental vCTB from which they originated. The ability to reprogram cells from term placenta into iTSC will allow study of early gestation events which impact placental function later in gestation, including preeclampsia and spontaneous preterm birth.
Keywords: Differentiation; Placenta; Reprogramming; Stem cell; Trophoblast.
Copyright © 2021 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Chu Cheung V, Peng CY, Marinic M, Sakabe NJ, Aneas I, Lynch VJ, Ober C, Nobrega MA, Kessler JA, 2021. Pluripotent cell cell-derived endometrial stromal fibroblasts in a cyclic, hormone-responsive, coculture model of human decidua. Cell Rep. 35(7):109138. - PubMed

