Associations between the gut microbiome and metabolome in early life
- PMID: 34454437
- PMCID: PMC8400760
- DOI: 10.1186/s12866-021-02282-3
Associations between the gut microbiome and metabolome in early life
Abstract
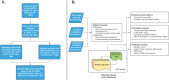
Background: The infant intestinal microbiome plays an important role in metabolism and immune development with impacts on lifelong health. The linkage between the taxonomic composition of the microbiome and its metabolic phenotype is undefined and complicated by redundancies in the taxon-function relationship within microbial communities. To inform a more mechanistic understanding of the relationship between the microbiome and health, we performed an integrative statistical and machine learning-based analysis of microbe taxonomic structure and metabolic function in order to characterize the taxa-function relationship in early life.
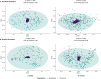
Results: Stool samples collected from infants enrolled in the New Hampshire Birth Cohort Study (NHBCS) at approximately 6-weeks (n = 158) and 12-months (n = 282) of age were profiled using targeted and untargeted nuclear magnetic resonance (NMR) spectroscopy as well as DNA sequencing of the V4-V5 hypervariable region from the bacterial 16S rRNA gene. There was significant inter-omic concordance based on Procrustes analysis (6 weeks: p = 0.056; 12 months: p = 0.001), however this association was no longer significant when accounting for phylogenetic relationships using generalized UniFrac distance metric (6 weeks: p = 0.376; 12 months: p = 0.069). Sparse canonical correlation analysis showed significant correlation, as well as identifying sets of microbe/metabolites driving microbiome-metabolome relatedness. Performance of machine learning models varied across different metabolites, with support vector machines (radial basis function kernel) being the consistently top ranked model. However, predictive R2 values demonstrated poor predictive performance across all models assessed (avg: - 5.06% -- 6 weeks; - 3.7% -- 12 months). Conversely, the Spearman correlation metric was higher (avg: 0.344-6 weeks; 0.265-12 months). This demonstrated that taxonomic relative abundance was not predictive of metabolite concentrations.
Conclusions: Our results suggest a degree of overall association between taxonomic profiles and metabolite concentrations. However, lack of predictive capacity for stool metabolic signatures reflects, in part, the possible role of functional redundancy in defining the taxa-function relationship in early life as well as the bidirectional nature of the microbiome-metabolome association. Our results provide evidence in favor of a multi-omic approach for microbiome studies, especially those focused on health outcomes.
Keywords: Functional redundancy; Infant gut microbiome; Metabolism; Prediction models; Stool metabolome.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures



References
-
- Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen A-M, Peet A, Tillmann V, Pöhö P, Mattila I, Lähdesmäki H, Franzosa EA, Vaarala O, de Goffau M, Harmsen H, Ilonen J, Virtanen SM, Clish CB, Orešič M, Huttenhower C, Knip M, DIABIMMUNE Study Group. Xavier RJ. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17(2):260–273. doi: 10.1016/j.chom.2015.01.001. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

