Nomograms predict survival of patients with lymph node-positive, luminal a breast cancer
- PMID: 34454451
- PMCID: PMC8401066
- DOI: 10.1186/s12885-021-08642-6
Nomograms predict survival of patients with lymph node-positive, luminal a breast cancer
Abstract
Background: To develop nomograms for the prediction of the 1-, 3-, and 5-year overall survival (OS) and breast cancer-specific survival (BCSS) for patients with lymph node positive, luminal A breast cancer.
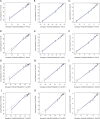
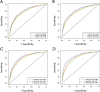
Methods: Thirty-nine thousand fifty-one patients from The Surveillance, Epidemiology, and End Results (SEER) database were included in our study and were set into a training group (n = 19,526) and a validation group (n = 19,525). Univariate analysis and Cox proportional hazards analysis were used to select variables and set up nomogram models on the basis of the training group. Kaplan-Meier curves and the log-rank test were adopted in the survival analysis and curves plotting. C-index, calibration plots and ROC curves were used to performed internal and external validation on the training group and validation group.
Results: Following independent factors were included in our nomograms: Age, marital status, grade, ethnic group, T stage, positive lymph nodes numbers, Metastasis, surgery, radiotherapy, chemotherapy. In both the training group and testing group, the calibration plots show that the actual and nomogram-predicted survival probabilities are consistent greatly. The C-index values of the nomograms in the training and validation cohorts were 0.782 and 0.806 for OS and 0.783 and 0.804 for BCSS, respectively. The ROC curves show that our nomograms have good discrimination.
Conclusions: The nomograms may assist clinicians predict the 1-, 3-, and 5-year OS and BCSS of patients with lymph node positive, luminal A breast cancer.
Keywords: Luminal A; Lymph node-positive; Nomograms; Prognosis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Nomograms to estimate long-term overall survival and breast cancer-specific survival of patients with luminal breast cancer.Oncotarget. 2016 Apr 12;7(15):20496-506. doi: 10.18632/oncotarget.7975. Oncotarget. 2016. PMID: 26967253 Free PMC article.
-
A novel nomogram and survival analysis for different lymph node status in breast cancer based on the SEER database.Breast Cancer. 2024 Sep;31(5):769-786. doi: 10.1007/s12282-024-01591-5. Epub 2024 May 27. Breast Cancer. 2024. PMID: 38802681 Free PMC article.
-
Nomogram for Predicting the Overall Survival of Patients With Breast Cancer With Pathologic Nodal Status N3.Clin Breast Cancer. 2020 Dec;20(6):e778-e785. doi: 10.1016/j.clbc.2020.06.002. Epub 2020 Jun 7. Clin Breast Cancer. 2020. PMID: 32636150
-
Multicentric and multifocal versus unifocal breast cancer: is the tumor-node-metastasis classification justified?Breast Cancer Res Treat. 2010 Jul;122(1):27-34. doi: 10.1007/s10549-010-0917-9. Epub 2010 May 8. Breast Cancer Res Treat. 2010. PMID: 20454925 Review.
-
Prognostic nomograms for young breast cancer: A retrospective study based on the SEER and METABRIC databases.Cancer Innov. 2024 Oct 25;3(6):e152. doi: 10.1002/cai2.152. eCollection 2024 Dec. Cancer Innov. 2024. PMID: 39464427 Free PMC article. Review.
Cited by
-
ERCC1 is a potential biomarker for predicting prognosis, immunotherapy, chemotherapy efficacy, and expression validation in HER2 over-expressing breast cancer.Front Oncol. 2022 Oct 20;12:955719. doi: 10.3389/fonc.2022.955719. eCollection 2022. Front Oncol. 2022. PMID: 36338712 Free PMC article.
-
A Nomogram for Predicting Liver Metastasis of Lymph-Node Positive Luminal B HER2 Negative Subtype Breast Cancer by Analyzing the Clinicopathological Characteristics of Patients with Breast Cancer.Technol Cancer Res Treat. 2022 Jan-Dec;21:15330338221132669. doi: 10.1177/15330338221132669. Technol Cancer Res Treat. 2022. PMID: 36254567 Free PMC article.
-
Constructing a neural network model based on tumor-infiltrating lymphocytes (TILs) to predict the survival of hepatocellular carcinoma patients.PeerJ. 2025 Apr 23;13:e19351. doi: 10.7717/peerj.19351. eCollection 2025. PeerJ. 2025. PMID: 40292102 Free PMC article.
-
Lymph Node Reporting and Data System (LN-RADS)-Retrospective Evaluation for Ultrasound Classification of Superficial Lymph Nodes.Cancers (Basel). 2025 Jun 18;17(12):2030. doi: 10.3390/cancers17122030. Cancers (Basel). 2025. PMID: 40563680 Free PMC article.
-
The clinical prognostic factors of patients with stage IB lung adenocarcinoma.Transl Cancer Res. 2021 Nov;10(11):4727-4738. doi: 10.21037/tcr-21-1174. Transl Cancer Res. 2021. PMID: 35116327 Free PMC article.
References
-
- Kosvyra A, Maramis C, Chouvarda I. Developing an Integrated Genomic Profile for Cancer Patients with the Use of NGS Data. Emerg Sci J. 2019;3(3):157–167. doi: 10.28991/esj-2019-01178. - DOI
-
- Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncology. 2011;22(8):1736–1747. doi: 10.1093/annonc/mdr304. - DOI - PMC - PubMed
-
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS et al: Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America 2001, 98(19):10869–10874.10.1073/pnas.191367098 - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

