CIDO ontology updates and secondary analysis of host responses to COVID-19 infection based on ImmPort reports and literature
- PMID: 34454610
- PMCID: PMC8400831
- DOI: 10.1186/s13326-021-00250-4
CIDO ontology updates and secondary analysis of host responses to COVID-19 infection based on ImmPort reports and literature
Abstract
Background: With COVID-19 still in its pandemic stage, extensive research has generated increasing amounts of data and knowledge. As many studies are published within a short span of time, we often lose an integrative and comprehensive picture of host-coronavirus interaction (HCI) mechanisms. As of early April 2021, the ImmPort database has stored 7 studies (with 6 having details) that cover topics including molecular immune signatures, epitopes, and sex differences in terms of mortality in COVID-19 patients. The Coronavirus Infectious Disease Ontology (CIDO) represents basic HCI information. We hypothesize that the CIDO can be used as the platform to represent newly recorded information from ImmPort leading the reinforcement of CIDO.
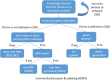
Methods: The CIDO was used as the semantic platform for logically modeling and representing newly identified knowledge reported in the 6 ImmPort studies. A recursive eXtensible Ontology Development (XOD) strategy was established to support the CIDO representation and enhancement. Secondary data analysis was also performed to analyze different aspects of the HCI from these ImmPort studies and other related literature reports.
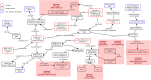
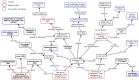
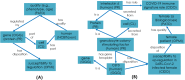
Results: The topics covered by the 6 ImmPort papers were identified to overlap with existing CIDO representation. SARS-CoV-2 viral S protein related HCI knowledge was emphasized for CIDO modeling, including its binding with ACE2, mutations causing different variants, and epitope homology by comparison with other coronavirus S proteins. Different types of cytokine signatures were also identified and added to CIDO. Our secondary analysis of two cohort COVID-19 studies with cytokine panel detection found that a total of 11 cytokines were up-regulated in female patients after infection and 8 cytokines in male patients. These sex-specific gene responses were newly modeled and represented in CIDO. A new DL query was generated to demonstrate the benefits of such integrative ontology representation. Furthermore, IL-10 signaling pathway was found to be statistically significant for both male patients and female patients.
Conclusion: Using the recursive XOD strategy, six new ImmPort COVID-19 studies were systematically reviewed, the results were modeled and represented in CIDO, leading to the enhancement of CIDO. The enhanced ontology and further seconary analysis supported more comprehensive understanding of the molecular mechanism of host responses to COVID-19 infection.
Keywords: CIDO; COVID-19; Coronavirus infectious disease ontology; Host-coronavirus interaction; ImmPort; Metadata; OBO foundry; Ontology; SARS-CoV-2; Sex difference.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
A comprehensive update on CIDO: the community-based coronavirus infectious disease ontology.J Biomed Semantics. 2022 Oct 21;13(1):25. doi: 10.1186/s13326-022-00279-z. J Biomed Semantics. 2022. PMID: 36271389 Free PMC article.
-
Visual comprehension and orientation into the COVID-19 CIDO ontology.J Biomed Inform. 2021 Aug;120:103861. doi: 10.1016/j.jbi.2021.103861. Epub 2021 Jul 2. J Biomed Inform. 2021. PMID: 34224898 Free PMC article.
-
The Infectious Disease Ontology in the age of COVID-19.J Biomed Semantics. 2021 Jul 18;12(1):13. doi: 10.1186/s13326-021-00245-1. J Biomed Semantics. 2021. PMID: 34275487 Free PMC article.
-
The eXtensible ontology development (XOD) principles and tool implementation to support ontology interoperability.J Biomed Semantics. 2018 Jan 12;9(1):3. doi: 10.1186/s13326-017-0169-2. J Biomed Semantics. 2018. PMID: 29329592 Free PMC article. Review.
-
Coronavirus disease 2019 and asthma, allergic rhinitis: molecular mechanisms and host-environmental interactions.Curr Opin Allergy Clin Immunol. 2021 Feb 1;21(1):1-7. doi: 10.1097/ACI.0000000000000699. Curr Opin Allergy Clin Immunol. 2021. PMID: 33186186 Review.
Cited by
-
Ontology-based taxonomical analysis of experimentally verified natural and laboratory human coronavirus hosts and its implication for COVID-19 virus origination and transmission.PLoS One. 2024 Jan 22;19(1):e0295541. doi: 10.1371/journal.pone.0295541. eCollection 2024. PLoS One. 2024. PMID: 38252647 Free PMC article.
-
Big knowledge visualization of the COVID-19 CIDO ontology evolution.BMC Med Inform Decis Mak. 2023 May 9;23(Suppl 1):88. doi: 10.1186/s12911-023-02184-6. BMC Med Inform Decis Mak. 2023. PMID: 37161560 Free PMC article.
-
Enriching the FIDEO ontology with food-drug interactions from online knowledge sources.J Biomed Semantics. 2024 Mar 4;15(1):1. doi: 10.1186/s13326-024-00302-5. J Biomed Semantics. 2024. PMID: 38438913 Free PMC article.
-
A new framework for host-pathogen interaction research.Front Immunol. 2022 Dec 15;13:1066733. doi: 10.3389/fimmu.2022.1066733. eCollection 2022. Front Immunol. 2022. PMID: 36591248 Free PMC article.
-
Immune Biomarkers, Profiles, and Responses: A Vaccine Ontology Perspective.bioRxiv [Preprint]. 2025 Jul 22:2025.07.18.665557. doi: 10.1101/2025.07.18.665557. bioRxiv. 2025. PMID: 40777277 Free PMC article. Preprint.
References
-
- CDC COVID-19 data tracker weekly review [https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html].
-
- Bhattacharya S, Dunn P, Thomas CG, Smith B, Schaefer H, Chen J, Hu Z, Zalocusky KA, Shankar RD, Shen-Orr SS, Thomson E, Wiser J, Butte AJ. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5(1):180015. doi: 10.1038/sdata.2018.15. - DOI - PMC - PubMed
-
- He Y, Yu H, Ong E, Wang Y, Liu Y, Huffman A, Huang HH, Beverley J, Hur J, Yang X, Chen L, Omenn GS, Athey B, Smith B. CIDO, a community-based ontology for coronavirus disease knowledge and data integration, sharing, and analysis. Sci Data. 2020;7(1):181. doi: 10.1038/s41597-020-0523-6. - DOI - PMC - PubMed
-
- Ribeiro MM, Wassermann R, Flouris G, Antoniou G. Minimal change: relevance and recovery revisited. Artif Intell. 2013;201:59–80. doi: 10.1016/j.artint.2013.06.001. - DOI
-
- Solimando A, Guerrini G. Ontology adaptation upon updates. In: Extended semantic web conference. v. 7955. Springer; 2013. p. 34–45. https://link.springer.com/chapter/10.1007/978-3-642-41242-4_4. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

