Relevance of immune response and vaccination strategies of SARS-CoV-2 in the phase of viral red queen dynamics
- PMID: 34454775
- PMCID: PMC8387243
- DOI: 10.1016/j.ijmmb.2021.08.001
Relevance of immune response and vaccination strategies of SARS-CoV-2 in the phase of viral red queen dynamics
Abstract
Background: Following a relatively mild first wave of coronavirus disease 2019 (COVID-19) in India, a deadly second wave of the pandemic overwhelmed the healthcare system due to the emergence of fast-transmitting SARS-CoV-2 genetic variants. The emergence and spread of the B.1.617.2/Delta variant considered to be driving the devastating second wave of COVID-19 in India. Currently, the Delta variant has rapidly overtaken the previously circulating variants to become the dominant strain. Critical mutations in the spike/RBD region of these variants have raised serious concerns about the virus's increased transmissibility and decreased vaccine effectiveness. As a result, significant scientific and public concern has been expressed about the impact of virus variants on COVID-19 vaccines.
Objectives: The purpose of this article is to provide an additional explanation in the context of the evolutionary trajectory of SARS-CoV-2 variants in India, the vaccine-induced immune response to the variants of concern (VOC), and various vaccine deployment strategies to rapidly increase population immunity.
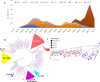
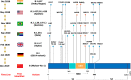
Content: Phylogenetic analysis of SARS-CoV-2 isolates circulating in India suggests the emergence and spread of B.1.617 variant. The immunogenicity of currently approved vaccines indicates that the majority of vaccines elicit an antibody response and some level of protection. According to current data, vaccines in the pre-fusion configuration (2p substitution) have an advantage in terms of nAb titer, but the duration of vaccine-induced immunity, as well as the role of T cells and memory B cells in protection, remain unknown. Since vaccine efficacy on virus variants is one of the major factors to be considered for achieving herd immunity, existing vaccines need to be improved or effective next-generation vaccines should be developed to cover the new variants of the virus.
Keywords: B.1.617; COVID-19; SARS-CoV-2; Variants.
Copyright © 2021 Indian Association of Medical Microbiologists. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Tegally H., Wilkinson E., Giovanetti M., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 Dec;20248640 doi: 10.1101/2020.12.21.20248640. - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

