Exosomal microRNA-15a from mesenchymal stem cells impedes hepatocellular carcinoma progression via downregulation of SALL4
- PMID: 34455417
- PMCID: PMC8403170
- DOI: 10.1038/s41420-021-00611-z
Exosomal microRNA-15a from mesenchymal stem cells impedes hepatocellular carcinoma progression via downregulation of SALL4
Abstract
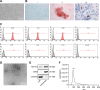
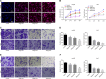
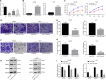
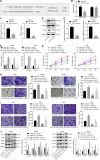
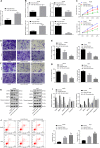
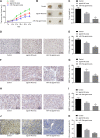
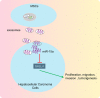
Hepatocellular carcinoma (HCC) is a heterogeneous tumor with an increased incidence worldwide accompanied by high mortality and dismal prognosis. Emerging evidence indicates that mesenchymal stem cells (MSCs)-derived exosomes possess protective effects against various human diseases by transporting microRNAs (miRNAs or miRs). We aimed to explore the role of exosomal miR-15a derived from MSCs and its related mechanisms in HCC. Exosomes were isolated from transduced MSCs and co-incubated with Hep3B and Huh7 cells. miR-15a expression was examined by RT-qPCR in HCC cells, MSCs, and secreted exosomes. CCK-8, transwell, and flow cytometry were used to detect the effects of miR-15a or spalt-like transcription factor 4 (SALL4) on cell proliferative, migrating, invasive, and apoptotic properties. A dual-luciferase reporter gene assay was performed to validate the predicted targeting relationship of miR-15a with SALL4. Finally, in vivo experiments in nude mice were implemented to assess the impact of exosome-delivered miR-15a on HCC. The exosomes from MSCs restrained HCC cell proliferative, migrating, and invasive potentials, and accelerated their apoptosis. miR-15a was expressed at low levels in HCC cells and could bind to SALL4, thus curtailing the proliferative, migrating, and invasive abilities of HCC cells. Exosomes successfully delivered miR-15a to HCC cells. Exosomal miR-15a depressed tumorigenicity and metastasis of HCC tumors in vivo. Overall, exosomal miR-15a from MSCs can downregulate SALL4 expression and thereby retard HCC development.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Emerging role of exosomal microRNA in liver cancer in the era of precision medicine; potential and challenges.Front Mol Biosci. 2024 Jun 27;11:1381789. doi: 10.3389/fmolb.2024.1381789. eCollection 2024. Front Mol Biosci. 2024. PMID: 38993840 Free PMC article. Review.
-
Exosomal lncRNA H19 promotes the progression of hepatocellular carcinoma treated with Propofol via miR-520a-3p/LIMK1 axis.Cancer Med. 2020 Oct;9(19):7218-7230. doi: 10.1002/cam4.3313. Epub 2020 Aug 7. Cancer Med. 2020. PMID: 32767662 Free PMC article.
-
Human umbilical cord-mesenchymal stem cells-derived exosomes carrying microRNA-15a-5p possess therapeutic effects on Wilms tumor via regulating septin 2.Bioengineered. 2022 Mar;13(3):6136-6149. doi: 10.1080/21655979.2022.2037379. Bioengineered. 2022. PMID: 35200105 Free PMC article.
-
Overexpressed Tumor Suppressor Exosomal miR-15a-5p in Cancer Cells Inhibits PD1 Expression in CD8+T Cells and Suppresses the Hepatocellular Carcinoma Progression.Front Oncol. 2021 Mar 19;11:622263. doi: 10.3389/fonc.2021.622263. eCollection 2021. Front Oncol. 2021. Retraction in: Front Oncol. 2024 Mar 05;14:1388383. doi: 10.3389/fonc.2024.1388383. PMID: 33816255 Free PMC article. Retracted.
-
Exosomal microRNAs from mesenchymal stem/stromal cells: Biology and applications in neuroprotection.World J Stem Cells. 2021 Jul 26;13(7):776-794. doi: 10.4252/wjsc.v13.i7.776. World J Stem Cells. 2021. PMID: 34367477 Free PMC article. Review.
Cited by
-
SALL4 in gastrointestinal tract cancers: upstream and downstream regulatory mechanisms.Mol Med. 2024 Apr 8;30(1):46. doi: 10.1186/s10020-024-00812-z. Mol Med. 2024. PMID: 38584262 Free PMC article. Review.
-
Exosomes Derived from Induced and Wharton's Jelly-Derived Mesenchymal Stem Cells Promote Senescence-like Features and Migration in Cancer Cells.Int J Mol Sci. 2025 Jun 26;26(13):6178. doi: 10.3390/ijms26136178. Int J Mol Sci. 2025. PMID: 40649957 Free PMC article.
-
Emerging functions and clinical applications of exosomal microRNAs in diseases.Noncoding RNA Res. 2023 May 9;8(3):350-362. doi: 10.1016/j.ncrna.2023.05.004. eCollection 2023 Sep. Noncoding RNA Res. 2023. PMID: 37250456 Free PMC article. Review.
-
Characterizing the pan-cancer role of exosomal miRNAs in metastasis across cancers.Comput Struct Biotechnol J. 2024 Dec 26;27:252-264. doi: 10.1016/j.csbj.2024.12.025. eCollection 2025. Comput Struct Biotechnol J. 2024. PMID: 39866667 Free PMC article.
-
Emerging role of exosomal microRNA in liver cancer in the era of precision medicine; potential and challenges.Front Mol Biosci. 2024 Jun 27;11:1381789. doi: 10.3389/fmolb.2024.1381789. eCollection 2024. Front Mol Biosci. 2024. PMID: 38993840 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

