Diverse methanogens, bacteria and tannase genes in the feces of the endangered volcano rabbit (Romerolagus diazi)
- PMID: 34458021
- PMCID: PMC8378336
- DOI: 10.7717/peerj.11942
Diverse methanogens, bacteria and tannase genes in the feces of the endangered volcano rabbit (Romerolagus diazi)
Abstract
Background: The volcano rabbit is the smallest lagomorph in Mexico, it is monotypic and endemic to the Trans-Mexican Volcanic Belt. It is classified as endangered by Mexican legislation and as critically endangered by the IUCN, in the Red List. Romerolagus diazi consumes large amounts of grasses, seedlings, shrubs, and trees. Pines and oaks contain tannins that can be toxic to the organisms which consume them. The volcano rabbit microbiota may be rich in bacteria capable of degrading fiber and phenolic compounds.
Methods: We obtained the fecal microbiome of three adults and one young rabbit collected in Coajomulco, Morelos, Mexico. Taxonomic assignments and gene annotation revealed the possible roles of different bacteria in the rabbit gut. We searched for sequences encoding tannase enzymes and enzymes associated with digestion of plant fibers such as cellulose and hemicellulose.
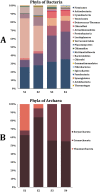
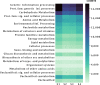
Results: The most representative phyla within the Bacteria domain were: Proteobacteria, Firmicutes and Actinobacteria for the young rabbit sample (S1) and adult rabbit sample (S2), which was the only sample not confirmed by sequencing to correspond to the volcano rabbit. Firmicutes, Actinobacteria and Cyanobacteria were found in adult rabbit samples S3 and S4. The most abundant phylum within the Archaea domain was Euryarchaeota. The most abundant genera of the Bacteria domain were Lachnoclostridium (Firmicutes) and Acinetobacter (Proteobacteria), while Methanosarcina predominated from the Archaea. In addition, the potential functions of metagenomic sequences were identified, which include carbohydrate and amino acid metabolism. We obtained genes encoding enzymes for plant fiber degradation such as endo 1,4 β-xylanases, arabinofuranosidases, endoglucanases and β-glucosidases. We also found 18 bacterial tannase sequences.
Keywords: Fecal metagenome; Herbivorous animals; Microbiome; Tannase enzymes; Volcano rabbit.
©2021 Montes-Carreto et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures




Similar articles
-
The complete mitochondrial genome of the 'Zacatuche' Volcano rabbit (Romerolagus diazi), an endemic and endangered species from the Volcanic Belt of Central Mexico.Mol Biol Rep. 2022 Feb;49(2):1141-1149. doi: 10.1007/s11033-021-06940-7. Epub 2021 Nov 16. Mol Biol Rep. 2022. PMID: 34783988
-
Response of the subalpine bunchgrasses to wildfires and its effects in the relative abundance of the volcano rabbit in the Ajusco-Chichinautzin Mountain Range.PeerJ. 2024 Jun 28;12:e17510. doi: 10.7717/peerj.17510. eCollection 2024. PeerJ. 2024. PMID: 38952973 Free PMC article.
-
Mining biomass-degrading genes through Illumina-based de novo sequencing and metagenomic analysis of free-living bacteria in the gut of the lower termite Coptotermes gestroi harvested in Vietnam.J Biosci Bioeng. 2014 Dec;118(6):665-71. doi: 10.1016/j.jbiosc.2014.05.010. Epub 2014 Jun 11. J Biosci Bioeng. 2014. PMID: 24928651
-
Metagenomic analysis of the Rhinopithecus bieti fecal microbiome reveals a broad diversity of bacterial and glycoside hydrolase profiles related to lignocellulose degradation.BMC Genomics. 2015 Mar 12;16(1):174. doi: 10.1186/s12864-015-1378-7. BMC Genomics. 2015. PMID: 25887697 Free PMC article.
-
Metagenomic insights into the roles of Proteobacteria in the gastrointestinal microbiomes of healthy dogs and cats.Microbiologyopen. 2018 Oct;7(5):e00677. doi: 10.1002/mbo3.677. Epub 2018 Jun 17. Microbiologyopen. 2018. PMID: 29911322 Free PMC article. Review.
Cited by
-
Short Term Effect of Ivermectin on the Bacterial Microbiota from Fecal Samples in Chinchillas (Chinchilla lanigera).Vet Sci. 2023 Feb 20;10(2):169. doi: 10.3390/vetsci10020169. Vet Sci. 2023. PMID: 36851473 Free PMC article.
-
The fecal microbiota of the mouse-eared bat (Myotis velifer) with new records of microbial taxa for bats.PLoS One. 2024 Dec 5;19(12):e0314847. doi: 10.1371/journal.pone.0314847. eCollection 2024. PLoS One. 2024. PMID: 39637086 Free PMC article.
-
Design and application of an efficient cellulose-degrading microbial consortium and carboxymethyl cellulase production optimization.Front Microbiol. 2022 Jul 15;13:957444. doi: 10.3389/fmicb.2022.957444. eCollection 2022. Front Microbiol. 2022. PMID: 35910619 Free PMC article.
References
-
- Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. 2013;5:627–640. doi: 10.3390/d5030627. - DOI
LinkOut - more resources
Full Text Sources

